Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/brainnetuoa/data_driven_network_neuroscience
https://github.com/brainnetuoa/data_driven_network_neuroscience
Last synced: 3 months ago
JSON representation
- Host: GitHub
- URL: https://github.com/brainnetuoa/data_driven_network_neuroscience
- Owner: brainnetuoa
- Created: 2023-09-24T23:58:21.000Z (9 months ago)
- Default Branch: main
- Last Pushed: 2023-10-18T22:30:17.000Z (8 months ago)
- Last Synced: 2024-01-17T12:03:08.535Z (5 months ago)
- Language: Python
- Size: 892 KB
- Stars: 6
- Watchers: 3
- Forks: 0
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
Lists
- awesome-stars - data_driven_network_neuroscience
README
[](https://github.com/psf/black)
# Data-Driven Network Neuroscience: On Data Collection and Benchmark
This repository contains a package of scripts and codes used in the paper to convert raw functional images to connectivity matrices using [fMRIPrep](https://fmriprep.org/en/stable/). The detailed description can be found in [our paper](https://arxiv.org/abs/2211.12421) on NeurIPS, Datasets and Benchmarks Track, 2023. The dataset we used and the preprocessed data is collected at [https://doi.org/10.17608/k6.auckland.21397377](https://doi.org/10.17608/k6.auckland.21397377).
## Samples of the raw MRI / preprocessed outputs / matrices
* A sample TaoWu subject can be accessed here: [sub-control032057-raw-image](https://auckland.figshare.com/articles/dataset/NeurIPS_2022_Datasets/21397377?file=42753967)
* The fMRIPrep outputs of the same subject fully preprocessed can be accessed here: [sub-control032057-preprocessed](https://auckland.figshare.com/articles/dataset/NeurIPS_2022_Datasets/21397377?file=42753970)
* The set of connectivity (using correlation) and time-series matrices generated for the same subject can be accessed here: [sub-control032057-matrices](https://auckland.figshare.com/articles/dataset/NeurIPS_2022_Datasets/21397377?file=42753964)
## Requirements
* Containerized execution environment:
* [Docker](https://www.nipreps.org/apps/docker/) or
* [Singularity](https://www.nipreps.org/apps/singularity/)
## External Dependencies
* [fMRIPrep](https://fmriprep.org/en/stable/index.html) (version 20.2.3)
* [dcm2niix](https://github.com/rordenlab/dcm2niix)
* [nilearn](https://nilearn.github.io/stable/index.html)
## Setup
1. Install numpy, os, shutil, glob, dcm2niix, nilearn, scipy modules for python programming
2. Install Docker/Singularity and fMRIPrep
* Installing fMRIPrep requires several steps and sorting out dependencies and a freesurfer license (free to acquire). We recommend following [this guide](https://andysbrainbook.readthedocs.io/en/latest/OpenScience/OS/fMRIPrep.html)
## Steps to preprocess neuroimages:
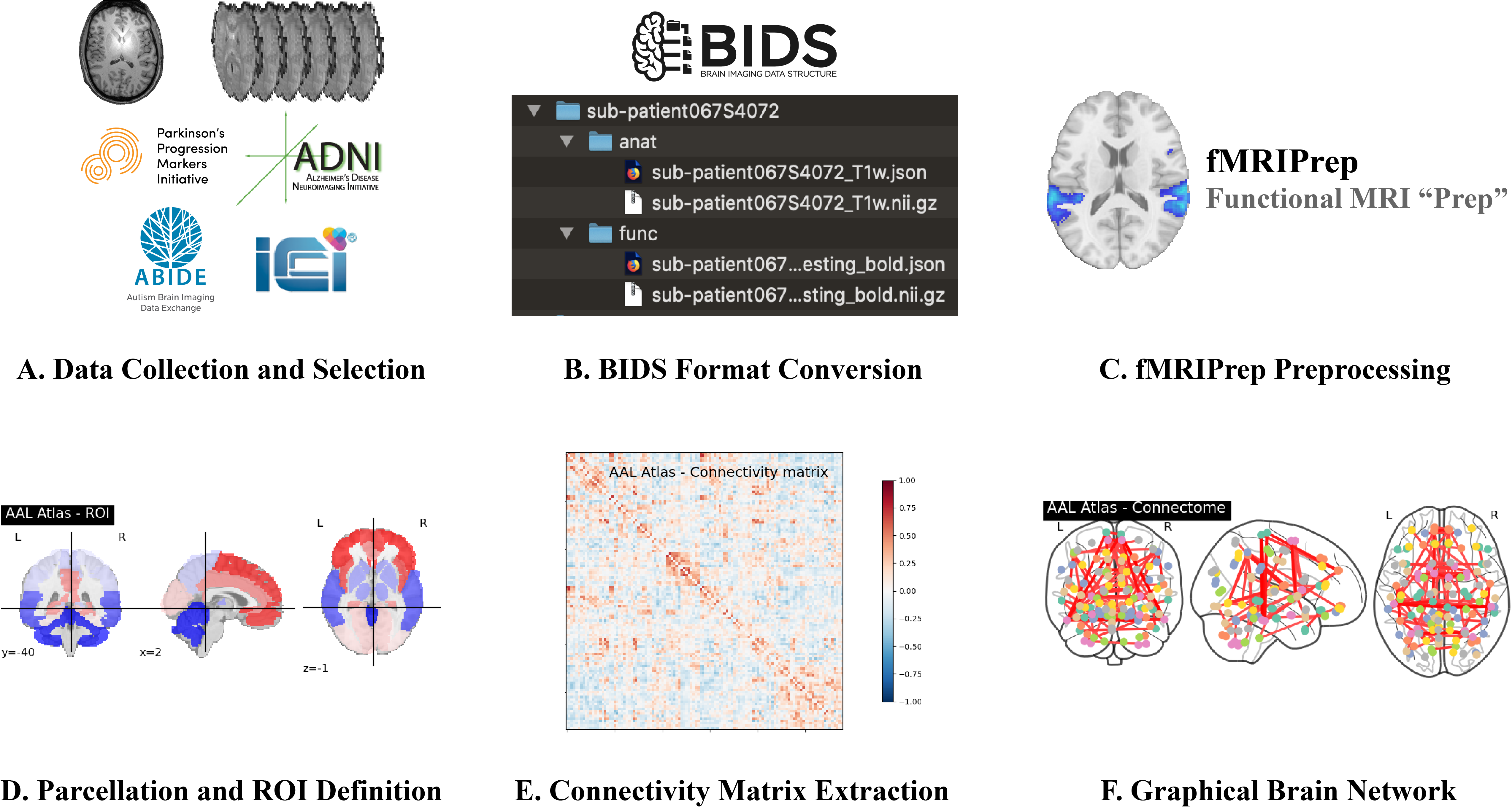
## Step A: Data Collection and Selection
Access Link for [Neurocon and TaoWu Dataset](http://fcon_1000.projects.nitrc.org/indi/retro/parkinsons.html).
Access Link for [ABIDE/ADNI/PPMI](https://ida.loni.usc.edu/login.jsp).
* A sample TaoWu subject can be accessed here: [sub-control032057-raw-image](https://auckland.figshare.com/articles/dataset/NeurIPS_2022_Datasets/21397377?file=42753967)
* Each fMRI image needs to be accompanied with a structural T1-weighted (T1w) image acquired from the same subject (ideally) in the same scan session
## Step B: BIDS Format Conversion
Raw T1w/fMRI data are in DICOM or NifTi format
* This step is to convert raw MRI data in either DICOM or NifTi into [BIDS format](https://bids.neuroimaging.io/)
* ABIDE_Nifti2BIDS.py - To convert raw MRI data (NifTi format) to BIDS - For ABIDE dataset
* ADNI_PPMI_DCM2BIDS.py - To convert raw MRI data (DICOM format) to BIDS - For PPMI and ADNI dataset
* Neurocon and TaoWu dataset are NifTi files and are already BIDS formatted
## Step C: fMRIPrep Preprocessing
Make sure you have installed fMRIPrep correctly using the information and guides from the links above.
* Preprocess BIDS formatted neuroimages (1 T1w image and 1 fMRI BOLD image) using fMRIPrep
* fmriprep_shellscript.sh - Script to execute fmriprep preprocessing
* fMRIPrep outputs a number of BIDS Derivative compliant files
* For details on each file: https://fmriprep.org/en/stable/outputs.html
A sample of a fully preprocessed TaoWu subject (and its outputs) can be accessed here: [sub-control032057-preprocessed](https://auckland.figshare.com/articles/dataset/NeurIPS_2022_Datasets/21397377?file=42753970)
## Steps D, E, and F: Parcellation and ROI Definition, Connectivity Matrix Extraction, and Graphical Brain Network
* Convert preprocessed Nifti images into connectivity matrices
* ConnectivityMatrices.py - Code to generate connectivity matrices
## Perform Experimental Analysis
* The codes we used to run our empirical analysis
* NIPS_paper_gridsearch_experiments.py
* EdgeWeightsStatistics.py
Note: Input/Output location and the required modification are detailed within the python codes
## Reference
```
@misc{xu2023datadriven,
title={Data-Driven Network Neuroscience: On Data Collection and Benchmark},
author={Jiaxing Xu and Yunhan Yang and David Tse Jung Huang and Sophi Shilpa Gururajapathy and Yiping Ke and Miao Qiao and Alan Wang and Haribalan Kumar and Josh McGeown and Eryn Kwon},
year={2023},
eprint={2211.12421},
archivePrefix={arXiv},
primaryClass={q-bio.NC}
}
```