Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/hdidx/hdidx
Approximate Nearest Neighbor (ANN) search for high-dimensional data.
https://github.com/hdidx/hdidx
Last synced: 3 months ago
JSON representation
Approximate Nearest Neighbor (ANN) search for high-dimensional data.
- Host: GitHub
- URL: https://github.com/hdidx/hdidx
- Owner: hdidx
- License: mit
- Created: 2014-11-07T15:15:23.000Z (over 9 years ago)
- Default Branch: master
- Last Pushed: 2018-09-03T00:56:59.000Z (almost 6 years ago)
- Last Synced: 2024-01-18T16:15:41.789Z (5 months ago)
- Language: Python
- Homepage: http://wanji.me/hdidx
- Size: 167 KB
- Stars: 90
- Watchers: 14
- Forks: 26
- Open Issues: 5
-
Metadata Files:
- Readme: README.md
- License: LICENSE.md
Lists
- my-awesome-awesomeness - hdidx - dimensional data, [hdidx-eval](https://github.com/hdidx/hdidx-eval/blob/master/eval_indexer.py). (Uncategorized / Uncategorized)
README
# **HDIdx**: Indexing High-Dimensional Data
[](https://pypi.python.org/pypi/hdidx/)
[](https://pypi.python.org/pypi/hdidx/)
[](https://raw.githubusercontent.com/wanji/hdidx/master/LICENSE.md)
## What is **HDIdx**?
**HDIdx** is a python package for approximate nearest neighbor (ANN) search. Nearest neighbor (NN) search is very challenging in high-dimensional space because of the [*Curse of Dimensionality*](https://en.wikipedia.org/wiki/Curse_of_dimensionality) problem. The basic idea of **HDIdx** is to compress the original feature vectors into compact binary codes, and perform approximate NN search instead of extract NN search. This can largely reduce the storage requirements and can significantly speed up the search.
## Architecture
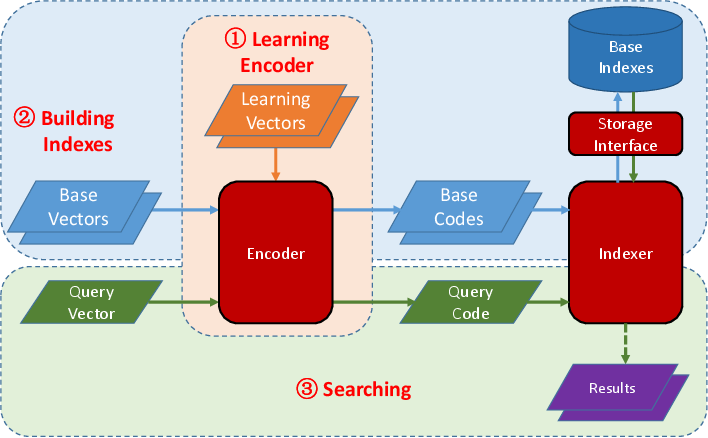
**HDIdx** has three main modules: 1) `Encoder` which can compress the original feature vectors into compact binary hash codes, 2) `Indexer` which can index the database items and search approximate nearest neighbor for a given query item, and 3) `Storage` module which encapsulates the underlying data storage, which can be memory or NoSQL database like LMDB, for the `Indexer`.
The current version implements following feature compressing algorithms:
- `Product Quantization`[1].
- `Spectral Hashing`[2].
To use HDIdx, first you should learn a `Encoder` from some learning vectors.
Then you can map the base vectors into hash codes using the learned `Encoder` and building indexes over these hash codes by an `Indexer`, which will write the indexes to the specified storage medium.
When a query vector comes, it will be mapped to hash codes by the same `Encoder` and the `Indexer` will find the similar items to this query vector.
## Installation
**HDIdx** can be installed by `pip`:
```bash
[sudo] pip install cython
[sudo] pip install hdidx
```
By default, **HDIdx** use kmeans algorithm provided by [*SciPy*](http://www.scipy.org/). To be more efficient, you can install python extensions of [*OpenCV*](http://opencv.org/), which can be installed via `apt-get` on Ubuntu. For other Linux distributions, e.g. CentOS, you need to compile it from source.
```bash
[sudo] apt-get install python-opencv
```
**HDIdx** will use [*OpenCV*](http://opencv.org/) automatically if it is available.
### Windows Guide
General dependencies:
- [Anaconda](https://store.continuum.io/cshop/anaconda/)
- [Microsoft Visual C++ Compiler for Python](http://www.microsoft.com/en-us/download/details.aspx?id=44266)
After install the above mentioned software, download [`stdint.h`](http://msinttypes.googlecode.com/svn/trunk/stdint.h) and put it under the `include` folder of Visual C++, e.g. `C:\Users\xxx\AppData\Local\Programs\Common\Microsoft\Visual C++ for Python\9.0\VC\include`. Then hdidx can be installed by `pip` from the *Anaconda Command Prompt*.
## Example
Here is a simple example. See this [notebook](http://nbviewer.ipython.org/gist/wanji/c08693f06ef744feef50) for more examples.
```python
# import necessary packages
import hdidx
import numpy as np
# generating sample data
ndim = 16 # dimension of features
ndb = 10000 # number of dababase items
nqry = 10 # number of queries
X_db = np.random.random((ndb, ndim))
X_qry = np.random.random((nqry, ndim))
# create Product Quantization Indexer
idx = hdidx.indexer.IVFPQIndexer()
# build indexer
idx.build({'vals': X_db, 'nsubq': 8})
# add database items to the indexer
idx.add(X_db)
# searching in the database, and return top-10 items for each query
ids, dis = idx.search(X_qry, 10)
print ids
print dis
```
## Citation
Please cite the following paper if you use this library:
```
@article{wan2015hdidx,
title={HDIdx: High-Dimensional Indexing for Efficient Approximate Nearest Neighbor Search},
author={Wan, Ji and Tang, Sheng and Zhang, Yongdong and Li, Jintao and Wu, Pengcheng and Hoi, Steven CH},
journal={Neurocomputing },
year={2016}
}
```
## Reference
```
[1] Jegou, Herve, Matthijs Douze, and Cordelia Schmid.
"Product quantization for nearest neighbor search."
Pattern Analysis and Machine Intelligence, IEEE Transactions on 33.1 (2011): 117-128.
[2] Weiss, Yair, Antonio Torralba, and Rob Fergus.
"Spectral hashing."
In Advances in neural information processing systems, pp. 1753-1760. 2009.
```