Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/JunMa11/AbdomenCT-1K
The official repository of "AbdomenCT-1K: Is Abdominal Organ Segmentation A Solved Problem?"
https://github.com/JunMa11/AbdomenCT-1K
Last synced: 3 months ago
JSON representation
The official repository of "AbdomenCT-1K: Is Abdominal Organ Segmentation A Solved Problem?"
- Host: GitHub
- URL: https://github.com/JunMa11/AbdomenCT-1K
- Owner: JunMa11
- License: apache-2.0
- Created: 2020-10-28T08:14:35.000Z (over 3 years ago)
- Default Branch: main
- Last Pushed: 2023-11-26T20:13:45.000Z (7 months ago)
- Last Synced: 2024-03-26T15:54:45.320Z (3 months ago)
- Language: Python
- Homepage: https://ieeexplore.ieee.org/document/9497733
- Size: 20 MB
- Stars: 188
- Watchers: 13
- Forks: 30
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Lists
- awesome_medical - AbdomenCT-1K
README
# AbdomenCT-1K [(Download)](https://forms.gle/dcqxBt4TeG8uTGvt7)
This is the official repository of "[AbdomenCT-1K](https://ieeexplore.ieee.org/document/9497733): Is Abdominal Organ Segmentation A Solved Problem?".
In this work,
- we collect a large and diverse abdominal CT organ segmentation dataset with 1000+ CT scans by augmenting the existing single organ datasets[1-6] with four abdominal organs, including liver, kidney, spleen, and pancreas.
- we conduct a large-scale study for liver, kidney, spleen, and pancreas segmentation and reveal the unsolved segmentation problems of the SOTA methods, such as the
limited generalization ability on distinct medical centers, phases, and unseen diseases.
- we further build four organ segmentation benchmarks for fully supervised, semi-supervised, weakly supervised, and continual learning, which are currently challenging and active research topics.
## News
- 2022: Welcome to join the MICCAI [FLARE 2022 Challenge](https://flare22.grand-challenge.org/): Fast and Low-resource semi-supervised Abdominal oRgan sEgmentation
- 2023: Welcome to join the MICCAI [FLARE 2023 Challenge](https://codalab.lisn.upsaclay.fr/competitions/12239): Fast, Low-resource, and Accurate oRgan and Pan-cancer sEgmentation
----------------
The whole AbdomenCT-1K dataset can be downloaded [here](https://forms.gle/XDrxSgoCXs3jzn8U7) (three parts) by filling in a simple data usage tracking form. The benchmark datasets are available on the `Dataset` page in the following grand-challenge homepages.
> Please register the challenges with your real names, affiliations, and affiliation E-mails.
## Fully Supervised Learning [(Grand-Challenge Homepage)](https://abdomenct-1k-fully-supervised-learning.grand-challenge.org/)

## Semi-supervised Learning [(Grand-Challenge Homepage)](https://abdomenct-1k-semi-supervised-learning.grand-challenge.org/)

## Weakly Supervised Learning [(Grand-Challenge Homepage)](https://abdomenct-1k-weaklysupervisedlearning.grand-challenge.org/)

## Continual Learning [(Grand-Challenge Homepage)](https://abdomenct-1k-continual-learning.grand-challenge.org/)

## Further Extentions
### 50 cases with 13 annotated organs (Download: [Zenodo](https://zenodo.org/record/7860267))
This dataset is used in [MICCAI FLARE2022](https://flare22.grand-challenge.org/) Challenge.
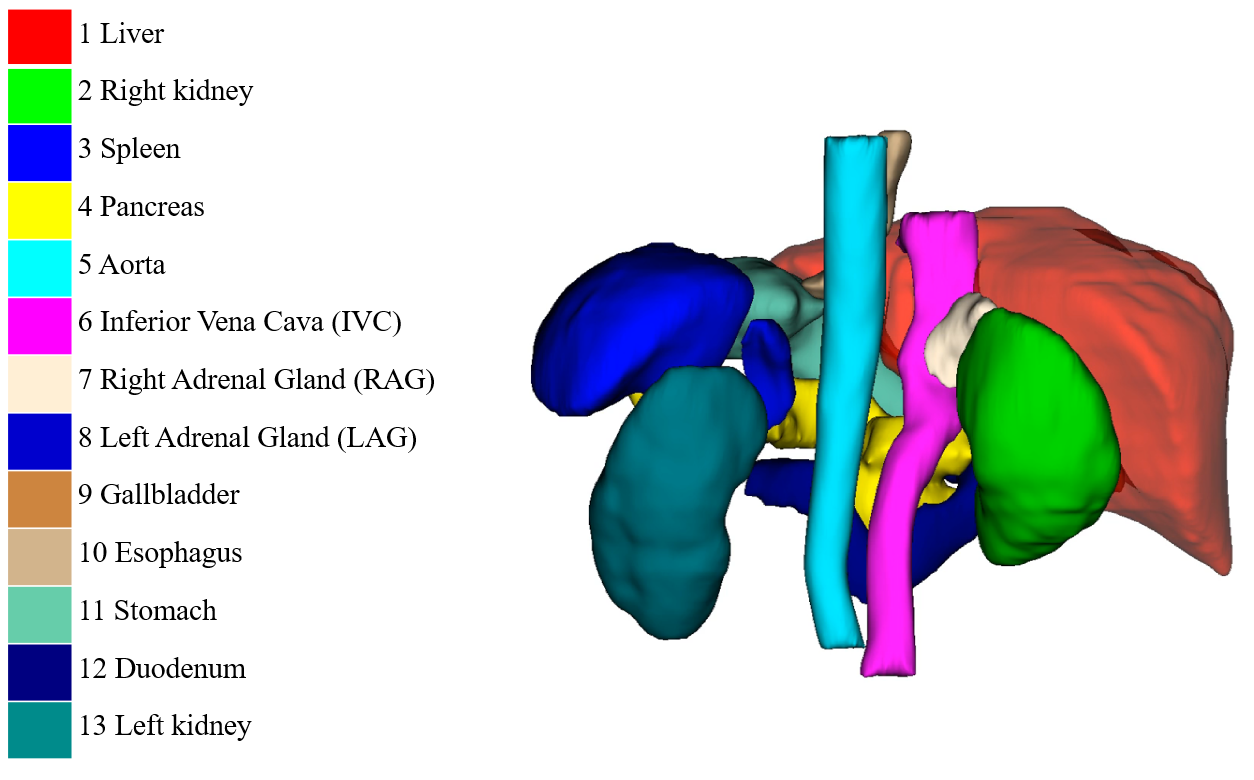
### 773 cases with pseudo tumor labels (Download: [Baidu Netdisk](https://pan.baidu.com/s/17FJfxAR6MVnYRiT-_dRNgA?pwd=2021), [Google Drive](https://drive.google.com/file/d/12AINDARYZDrdc66v891YT6VDxW9NsfjR/view?usp=sharing))
> Update: we add labels of 110 cases. Now, it contains 773 cases with pseudo tumor labels. The corresponding images are in the whole dataset `Case_00001-00773`.
It is challenging to make a definite and accurate diagnosis with only single phase CT scans because identifying the (malignant) tumor usually requires pathological examinations. As an alternative, we provide pseudo tumor labels of 663 cases by annotating all the possible tumors, which can be used for noisy label learning.

## References
[1] N. Heller, F. Isensee, K. H. Maier-Hein, X. Hou, C. Xie, F. Li, Y. Nan, G. Mu, Z. Lin, M. Han et al., “The state of the art in kidney and kidney tumor segmentation in contrast-enhanced ct imaging: Results of the kits19 challenge,” Medical Image Analysis, vol. 67, p. 101821, 2021.
[2] A. L. Simpson, M. Antonelli, S. Bakas, M. Bilello, K. Farahani, B. Van Ginneken, A. Kopp-Schneider, B. A. Landman, G. Litjens, B. Menze et al., “A large annotated medical image dataset for the development and evaluation of segmentation algorithms,” arXiv preprint arXiv:1902.09063, 2019.
[3] K. Clark, B. Vendt, K. Smith, J. Freymann, J. Kirby, P. Koppel, S. Moore, S. Phillips, D. Maffitt, M. Pringle et al., “The cancer imaging archive (tcia): maintaining and operating a public information repository,” Journal of Digital Imaging, vol. 26, no. 6, pp. 1045–1057, 2013.
[4] P. Bilic, P. F. Christ, E. Vorontsov, G. Chlebus, H. Chen, Q. Dou, C.-W. Fu, X. Han, P.-A. Heng, J. Hesser et al., "The liver tumor segmentation benchmark (lits)," arXiv preprint arXiv:1901.04056, 2019.
[5] H. R. Roth, A. Farag, E. B. Turkbey, L. Lu, J. Liu, and R. M. Summers, “Data from pancreas-CT,” The Cancer Imaging Archive, 2016.
[6] H. R. Roth, L. Lu, A. Farag, H.-C. Shin, J. Liu, E. B. Turkbey, and R. M. Summers, “Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation,” in International Conference on Medical Image Computing and Computer-assisted Intervention, 2015, pp. 556–564.
## Acknowledgment
We highly appreciate the organizers and contributors of [NIH Pancreas dataset](https://wiki.cancerimagingarchive.net/display/Public/Pancreas-CT), [Liver and Liver Tumor Segmentation challenge](https://competitions.codalab.org/competitions/15595), [Medical Segmentation Decathlon](http://medicaldecathlon.com/), and [Kidney Tumor Segmentation challenge (KiTS19)](https://kits19.grand-challenge.org/) for providing the publicly available abdominal CT datasets. We are grateful to the editors and the reviewers for their time and efforts spent on our paper. Their comments are very valuable for us to improve this work. We also thank the High Performance Computing Center of Nanjing University for supporting the blade cluster system to run the experiments. We also thank Mengzhang Li and Xiao Ma for helping us run some experiments.
## Citation
```
@article{Ma-2021-AbdomenCT-1K,
author={Ma, Jun and Zhang, Yao and Gu, Song and Zhu, Cheng and Ge, Cheng and Zhang, Yichi and An, Xingle and Wang, Congcong and Wang, Qiyuan and Liu, Xin and Cao, Shucheng and Zhang, Qi and Liu, Shangqing and Wang, Yunpeng and Li, Yuhui and He, Jian and Yang, Xiaoping},
journal={IEEE Transactions on Pattern Analysis and Machine Intelligence},
title={AbdomenCT-1K: Is Abdominal Organ Segmentation a Solved Problem?},
year={2022},
volume={44},
number={10},
pages={6695-6714},
doi={10.1109/TPAMI.2021.3100536}}
}
```