Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/krassowski/complex-upset
A library for creating complex UpSet plots with ggplot2 geoms
https://github.com/krassowski/complex-upset
ggplot ggplot2 patchwork python rstat rstats upset upsetr venn venn-diagram visualization
Last synced: 3 months ago
JSON representation
A library for creating complex UpSet plots with ggplot2 geoms
- Host: GitHub
- URL: https://github.com/krassowski/complex-upset
- Owner: krassowski
- License: mit
- Created: 2020-01-26T16:06:20.000Z (over 4 years ago)
- Default Branch: master
- Last Pushed: 2024-03-09T15:04:43.000Z (4 months ago)
- Last Synced: 2024-03-10T15:40:51.506Z (4 months ago)
- Topics: ggplot, ggplot2, patchwork, python, rstat, rstats, upset, upsetr, venn, venn-diagram, visualization
- Language: Jupyter Notebook
- Homepage:
- Size: 43.4 MB
- Stars: 417
- Watchers: 8
- Forks: 24
- Open Issues: 60
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Lists
- awesome-r-dataviz - complex-upset - A library for creating complex UpSet plots with ggplot2 geoms. (ggplot / Additional Plot Types)
- awesome-stars - krassowski/complex-upset - A library for creating complex UpSet plots with ggplot2 geoms (R)
README
# ComplexUpset
[](https://github.com/krassowski/complex-upset/actions?query=workflow%3Atest)
[](https://app.codecov.io/gh/krassowski/complex-upset)
[](https://CRAN.R-project.org/package=ComplexUpset)
[](https://anaconda.org/conda-forge/r-complexupset)
[](https://zenodo.org/badge/latestdoi/236336935)
Quick links: [Documentation](https://krassowski.github.io/complex-upset/) | [Installation](https://github.com/krassowski/complex-upset#Install) | [R examples](https://krassowski.github.io/complex-upset/articles/Examples_R.html) | [Python examples](https://krassowski.github.io/complex-upset/articles/Examples_Python.html)
## Showcase
Get all the goodies of `UpSetR`, but with full extensibility of `ggplot2`.
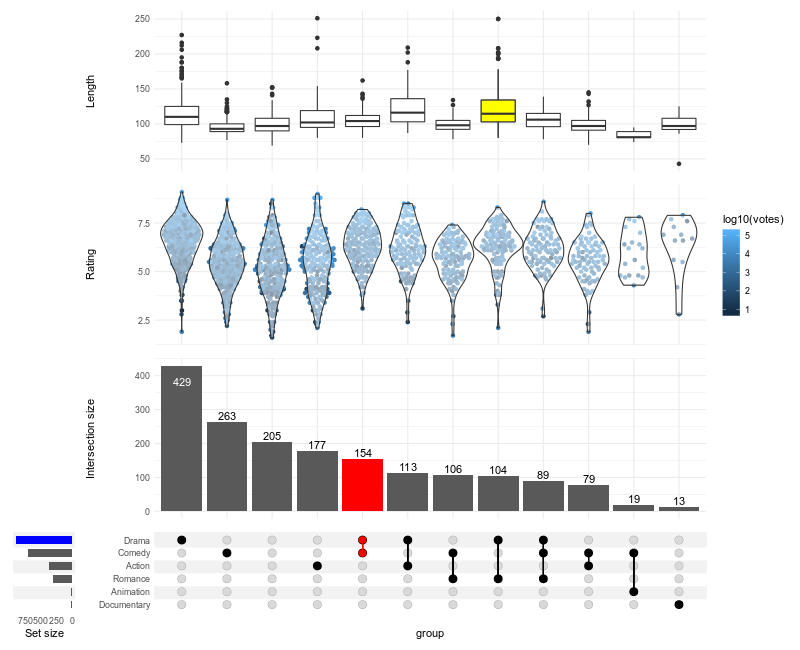
Click here to display the source code
```R
library(ggplot2)
library(ComplexUpset)
if(!require(ggplot2movies)) install.packages('ggplot2movies')
movies = ggplot2movies::movies
genres = c('Action', 'Animation', 'Comedy', 'Drama', 'Documentary', 'Romance')
upset(
movies,
genres,
annotations = list(
'Length'=ggplot(mapping=aes(x=intersection, y=length)) + geom_boxplot(),
'Rating'=ggplot(mapping=aes(x=intersection, y=rating))
# if you do not want to install ggbeeswarm, you can use geom_jitter
+ ggbeeswarm::geom_quasirandom(aes(color=log10(votes)))
+ geom_violin(width=1.1, alpha=0.5)
),
queries=list(
upset_query(
intersect=c('Drama', 'Comedy'),
color='red',
fill='red',
only_components=c('intersections_matrix', 'Intersection size')
),
upset_query(
set='Drama',
fill='blue'
),
upset_query(
intersect=c('Romance', 'Drama'),
fill='yellow',
only_components=c('Length')
)
),
min_size=10,
width_ratio=0.1
)
```
The full list of examples is available in the [documentation](https://krassowski.github.io/complex-upset/articles/Examples_Python.html); it also contains instructions for the use from Python.
## Install
To get the most recent version, open `R` and run:
```R
if(!require(devtools)) install.packages("devtools")
devtools::install_github("krassowski/complex-upset")
```
Alternatively, to get a stable CRAN release (which may be one version behind at times):
```R
install.packages('ComplexUpset')
```
Or, if you use conda/mamba:
```R
conda install -c conda-forge r-complexupset
```
## How it compares to other packages?
- [UpSetR](https://github.com/hms-dbmi/UpSetR) is a powerful tool and the pioneer in the UpSet visualisations; it was not designed to be extended with ggplot components. Unfortunately, the repository is no longer active (for two years now).
- [ggupset](https://github.com/const-ae/ggupset) uses scales to convert a single plot to upsets plots, e.g. with `scale_x_upset` - a really nice approach, recommended for simple ggplot.
- a cowplot-based upset was demonstrated in an [online publication](https://rpubs.com/alexeilutay/upsetr), however cowplot (a great tool at the time) got superseded by even greater tool: [patchwork](https://github.com/thomasp85/patchwork), which is what is used by ComplexUpset.
- [ComplexHeatmap](https://github.com/jokergoo/ComplexHeatmap) also offers a way to generate [UpSet plots with annotations](https://jokergoo.github.io/ComplexHeatmap-reference/book/upset-plot.html); while not ggplot2-centered, it provides extensive customization options, with a clear API. It may be the best choice if you already use it for heatmaps.
Use whichever tool you find the most useful for your particular use. The rational of making this repository public is not only to share the code, but also to demonstrate how simple it is now to create complex visualisations with patchwork (without the need to learn the ggplot internals).
For the interactive use, check out the [VCG/upset](https://github.com/VCG/upset). Regardless of the tool chosen, you may want to cite [(Lex et al, 2014)](https://dx.doi.org/10.1109/TVCG.2014.2346248) when using UpSet plots, especially in fields still dominated by Venn diagrams.
## Get inspired
Here are example publications and preprints including figures generated with this library:
- Martín-Martín et al. (2020). [Google Scholar, Microsoft Academic, Scopus, Dimensions, Web of Science, and OpenCitations’ COCI: a multidisciplinary comparison of coverage via citations](https://doi.org/10.1007/s11192-020-03690-4), *Scientometrics*: [Figure 2](https://link.springer.com/article/10.1007/s11192-020-03690-4#Fig2)
- Krassowski et al. (2020). [Analyses for "State of the field in multi-omics research: from computational needs to data mining and sharing](https://doi.org/10.3389/fgene.2020.610798), *Frontiers in Genetics*: [Figure 5](https://www.frontiersin.org/articles/10.3389/fgene.2020.610798/full#F5)
- Green et al. (2020). [Metabolic correlates of late midlife cognitive function: findings from the 1946 British Birth Cohort](https://doi.org/10.1101/2020.11.23.20236463), *medRxiv*: [Figure 2](https://www.medrxiv.org/content/10.1101/2020.11.23.20236463v2.full#F2)
- McDaniel et al. (2020). [Metabolic differentiation of co-occurring Accumulibacter clades revealed through genome-resolved metatranscriptomics](https://doi.org/10.1101/2020.11.23.394700), *bioRxiv* [Figure 3](https://www.biorxiv.org/content/10.1101/2020.11.23.394700v1.full#F3)
- Kozlowski et al. (2020). [Transposable Elements are an evolutionary force shaping genomic plasticity in the parthenogenetic root-knot nematode Meloidogyne incognita](https://doi.org/10.1101/2020.04.30.069948), *bioRxiv*: [Figure 7](https://www.biorxiv.org/content/10.1101/2020.04.30.069948v4.full#F7)
- Swamy et al. (2020). [A long read optimized de novo transcriptome pipeline reveals novel ocular developmentally regulated gene isoforms and disease targets](https://doi.org/10.1101/2020.08.21.261644), *bioRxiv*: [Figure 4](https://www.biorxiv.org/content/10.1101/2020.08.21.261644v2.full#F4)
The list is not meant to be exhaustive, but representative of applications to different fields, and of different usage ideas. You are welcome to add your own publication by [suggesting an edit](https://github.com/krassowski/complex-upset/edit/master/README.md).
## Testing
The unit tests are run with `testhat`, and the visual "doppelganger" tests use `vdiffr`.
The test cases for visual tests are auto-generated from the examples in the documentation; after changing or adding an example, please run:
- `scripts/prepare_for_tests.sh` to generate updated test cases, and
- `scripts/manage_visual_tests.sh` to validate the generated images.
## Acknowledgements
Originally developed in course of a DPhil programme in Women's & Reproductive Health at [OxfordNuffieldWRH](https://github.com/OxfordNuffieldWRH).