Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/EagleW/PaperRobot
Code for PaperRobot: Incremental Draft Generation of Scientific Ideas
https://github.com/EagleW/PaperRobot
attention-mechanism datasets end-to-end-learning generation memory-networks natural-language-generation nlp paper-generation pytorch text-generation
Last synced: 3 months ago
JSON representation
Code for PaperRobot: Incremental Draft Generation of Scientific Ideas
- Host: GitHub
- URL: https://github.com/EagleW/PaperRobot
- Owner: EagleW
- License: mit
- Created: 2019-05-18T02:03:32.000Z (about 5 years ago)
- Default Branch: master
- Last Pushed: 2021-05-31T03:49:51.000Z (about 3 years ago)
- Last Synced: 2024-01-07T10:54:32.985Z (6 months ago)
- Topics: attention-mechanism, datasets, end-to-end-learning, generation, memory-networks, natural-language-generation, nlp, paper-generation, pytorch, text-generation
- Language: Python
- Homepage: https://aclanthology.org/P19-1191
- Size: 63.8 MB
- Stars: 469
- Watchers: 46
- Forks: 135
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Lists
- awesome-nlg - PaperRobot: Incremental Draft Generation of Scientific Ideas - We present a PaperRobot who performs as an automatic research assistant. (Neural Natural Language Generation)
- awesome-scholarly-data-analysis - PaperRobot - includes PubMed Paper Reading Dataset
README
# PaperRobot: Incremental Draft Generation of Scientific Ideas
[PaperRobot: Incremental Draft Generation of Scientific Ideas](https://aclanthology.org//P19-1191)
[[Sample Output]](https://eaglew.github.io/PaperRobot/)
Accepted by 57th Annual Meeting of the Association for Computational Linguistics (ACL 2019)
Table of Contents
=================
* [Overview](#overview)
* [Requirements](#requirements)
* [Quickstart](#quickstart)
* [Citation](#citation)
## Overview
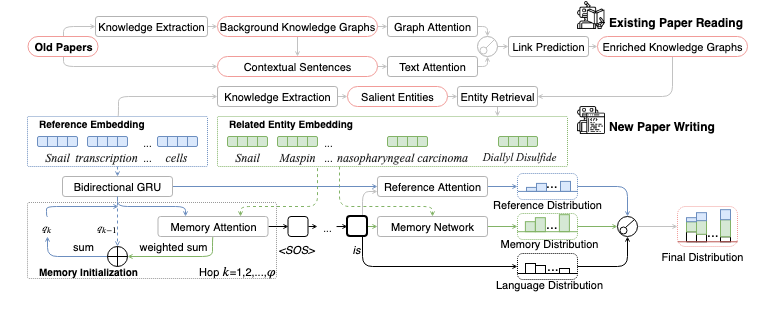
## Requirements
#### Environment:
- Python 3.6 **CAUTION!! The model might not be saved and loaded properly under Python 3.5**
- Ubuntu 16.04/18.04 **CAUTION!! The model might not run properly on windows because [windows uses backslashes on the path while Linux/OS X uses forward slashes](https://www.howtogeek.com/181774/why-windows-uses-backslashes-and-everything-else-uses-forward-slashes/)**
#### Pacakges
You can click the following links for detailed installation instructions.
- [Pytorch 1.1](https://pytorch.org/get-started/previous-versions/)
- [NumPy 1.16.3](https://www.scipy.org/install.html)
- [SciPy 1.2.1](https://www.scipy.org/install.html)
- [NetworkX 2.3](https://networkx.github.io/documentation/stable/install.html)
#### Data:
- [PubMed Paper Reading Dataset](https://drive.google.com/open?id=1DLmxK5x7m8bDPK5ZfAROtGpkWZ_v980Z)
This dataset gathers 14,857 entities, 133 relations, and entities corresponding tokenized text from PubMed. It contains 875,698 training pairs, 109,462 development pairs, and 109,462 test pairs.
- [PubMed Term, Abstract, Conclusion, Title Dataset](https://drive.google.com/open?id=1O91gX2maPHdIRUb9DdZmUOI5issRMXMY)
This dataset gathers three types of pairs: Title-to-Abstract (Training: 22,811/Development: 2095/Test: 2095), Abstract-to-Conclusion and Future work (Training: 22,811/Development: 2095/Test: 2095), Conclusion and Future work-to-Title (Training: 15,902/Development: 2095/Test: 2095) from PubMed. Each pair contains a pair of input and output as well as the corresponding terms(from original KB and link prediction results).
## Quickstart
### Existing paper reading
**CAUTION!! Because the dataset is quite large, the training and evaluation of link prediction model will be pretty slow.**
#### Preprocessing:
Download and unzip the `paper_reading.zip` from [PubMed Paper Reading Dataset](https://drive.google.com/open?id=1DLmxK5x7m8bDPK5ZfAROtGpkWZ_v980Z)
. Put `paper_reading` folder under the `Existing paper reading` folder.
#### Training
Hyperparameter can be adjusted as follows: For example, if you want to change the number of hidden unit to 6, you can append `--hidden 6` after `train.py`
```
python train.py
```
To resume training, you can apply the following command and put the previous model path after the `--model`
```
python train.py --cont --model models/GATA/best_dev_model.pth.tar
```
#### Test
Put the finished model path after the `--model`
The `test.py` will provide the ranking score for the test set.
```
python test.py --model models/GATA/best_dev_model.pth.tar
```
### New paper writing
#### Preprocessing:
Download and unzip the `data_pubmed_writing.zip` from [PubMed Term, Abstract, Conclusion, Title Dataset](https://drive.google.com/open?id=1O91gX2maPHdIRUb9DdZmUOI5issRMXMY)
. Put `data` folder under the `New paper writing folder`.
#### Training
Put the type of data after the `--data_path`. For example, if you want to train an abstract model, put `data/pubmed_abstract` after `--data_path`.
Put the model directory after the `--model_dp`
```
python train.py --data_path data/pubmed_abstract --model_dp abstract_model/
```
To resume training, you can apply the following command and put the previous model path after the `--model`
```
python train.py --data_path data/pubmed_abstract --cont --model abstract_model/memory/best_dev_model.pth.tar
```
For more other options, please check the code.
#### Test
Put the finished model path after the `--model`
The `test.py` will provide the score for the test set.
```
python test.py --data_path data/pubmed_abstract --model abstract_model/memory/best_dev_model.pth.tar
```
#### Predict an instance
Put the finished model path after the `--model`
The `input.py` will provide the prediction for customized input.
```
python input.py --data_path data/pubmed_abstract --model abstract_model/memory/best_dev_model.pth.tar
```
## Citation
```
@inproceedings{wang-etal-2019-paperrobot,
title = "{P}aper{R}obot: Incremental Draft Generation of Scientific Ideas",
author = "Wang, Qingyun and
Huang, Lifu and
Jiang, Zhiying and
Knight, Kevin and
Ji, Heng and
Bansal, Mohit and
Luan, Yi",
booktitle = "Proceedings of the 57th Conference of the Association for Computational Linguistics",
month = jul,
year = "2019",
address = "Florence, Italy",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/P19-1191",
pages = "1980--1991"
}
```
