https://github.com/AllenCellModeling/napari-aicsimageio
Multiple file format reading directly into napari using pure Python
https://github.com/AllenCellModeling/napari-aicsimageio
imageio microscopy napari
Last synced: 9 months ago
JSON representation
Multiple file format reading directly into napari using pure Python
- Host: GitHub
- URL: https://github.com/AllenCellModeling/napari-aicsimageio
- Owner: AllenCellModeling
- License: gpl-3.0
- Created: 2020-03-20T17:52:38.000Z (almost 6 years ago)
- Default Branch: main
- Last Pushed: 2024-07-21T00:01:26.000Z (over 1 year ago)
- Last Synced: 2025-04-14T02:38:54.584Z (10 months ago)
- Topics: imageio, microscopy, napari
- Language: Python
- Homepage:
- Size: 54.8 MB
- Stars: 35
- Watchers: 6
- Forks: 10
- Open Issues: 19
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
- Code of conduct: CODE_OF_CONDUCT.md
Awesome Lists containing this project
- awesome-biological-image-analysis - Napari-aicsimageio - Multiple file format reading directly into napari using pure Python. (Other)
README
# napari-aicsimageio
[](https://github.com/AllenCellModeling/napari-aicsimageio/raw/main/LICENSE)
[](https://github.com/AllenCellModeling/napari-aicsimageio/actions)
[](https://codecov.io/gh/AllenCellModeling/napari-aicsimageio)
AICSImageIO bindings for napari
---
## Features
- Supports reading metadata and imaging data for:
- `OME-TIFF`
- `TIFF`
- `CZI` (Zeiss)
- `LIF` (Leica)
- `ND2` (Nikon)
- `DV` (DeltaVision)
- Any formats supported by [aicsimageio](https://github.com/AllenCellModeling/aicsimageio)
- Any formats supported by [bioformats](https://github.com/tlambert03/bioformats_jar)
(Note: requires `java` and `mvn` executables)
- `SLD` (Slidebook)
- `SVS` (Aperio)
- [Full List](https://docs.openmicroscopy.org/bio-formats/6.5.1/supported-formats.html)
- Any additional format supported by [imageio](https://github.com/imageio/imageio)
- `PNG`
- `JPG`
- `GIF`
- `AVI`
- [Full List](https://imageio.readthedocs.io/en/v2.4.1/formats.html)
_While upstream `aicsimageio` is released under BSD-3 license, this plugin is released under GPLv3 license because it installs all format reader dependencies._
## Installation
**Stable Release:** `pip install napari-aicsimageio` or `conda install napari-aicsimageio -c conda-forge`
**Development Head:** `pip install git+https://github.com/AllenCellModeling/napari-aicsimageio.git`
> **Warning**
> The `bioformats` reader requires `java` and `mvn` executables, which cannot be pip installed.
> As a result, it's simplest to install it from conda-forge, ensuring both are also installed:
> `conda install -c conda-forge bioformats_jar`
### Reading Mode Threshold
This image reading plugin will load the provided image directly into memory if it meets
the following two conditions:
1. The filesize is less than 4GB.
2. The filesize is less than 30% of machine memory available.
If either of these conditions isn't met, the image is loaded in chunks only as needed.
### Use napari-aicsimageio as the Reader for All File Formats
If you want to force napari to always use this plugin as the reader for all file formats,
try running this snippet:
```python
from napari.settings import get_settings
get_settings().plugins.extension2reader = {'*': 'napari-aicsimageio', **get_settings().plugins.extension2reader}
```
For more details, see [#37](https://github.com/AllenCellModeling/napari-aicsimageio/issues/37).
## Examples of Features
#### General Image Reading
All image file formats supported by
[aicsimageio](https://github.com/AllenCellModeling/aicsimageio) will be read and all
raw data will be available in the napari viewer.
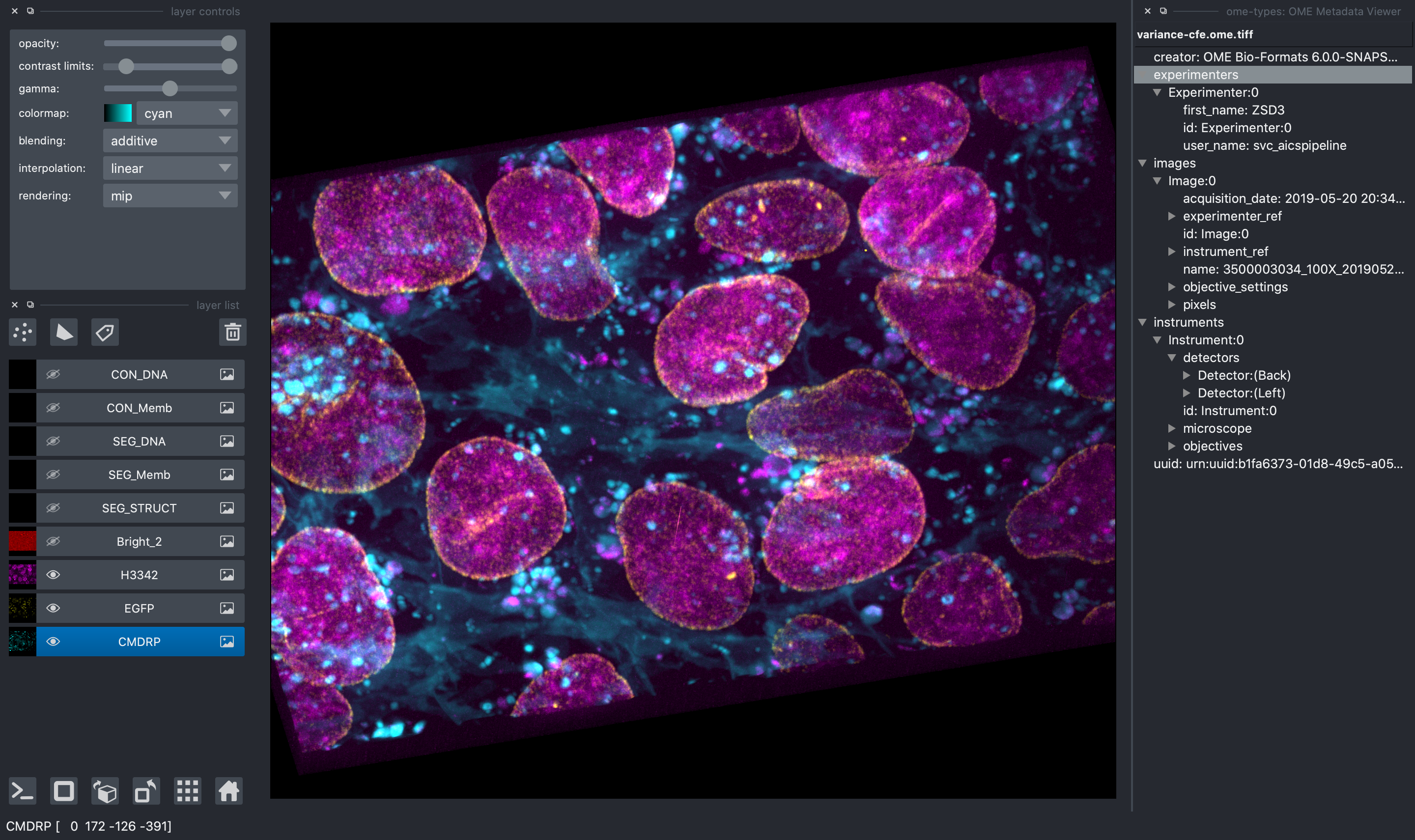
In addition, when reading an OME-TIFF, you can view all OME metadata directly in the
napari viewer thanks to `ome-types`.
#### Multi-Scene Selection
When reading a multi-scene file, a widget will be added to the napari viewer to manage
scene selection (clearing the viewer each time you change scene or adding the
scene content to the viewer) and a list of all scenes in the file.
#### Access to the AICSImage Object and Metadata
![napari viewer with console open showing `viewer.layers[0].metadata`](https://raw.githubusercontent.com/AllenCellModeling/napari-aicsimageio/main/images/console-access.png)
You can access the `AICSImage` object used to load the image pixel data and
image metadata using the built-in napari console:
```python
img = viewer.layers[0].metadata["aicsimage"]
img.dims.order # TCZYX
img.channel_names # ["Bright", "Struct", "Nuc", "Memb"]
img.get_image_dask_data("ZYX") # dask.array.Array
```
The napari layer metadata dictionary also stores a shorthand
for the raw image metadata:
```python
viewer.layers[0].metadata["raw_image_metadata"]
```
The metadata is returned in whichever format is used by the underlying
file format reader, i.e. for CZI the raw metadata is returned as
an `xml.etree.ElementTree.Element`, for OME-TIFF the raw metadata is returned
as an `OME` object from `ome-types`.
Lastly, if the underlying file format reader has an OME metadata conversion function,
you may additionally see a key in the napari layer metadata dictionary
called `"ome_types"`. For example, because the AICSImageIO
`CZIReader` and `BioformatsReader` both support converting raw image metadata
to OME metadata, you will see an `"ome_types"` key that stores the metadata transformed
into the OME metadata model.
```python
viewer.layers[0].metadata["ome_types"] # OME object from ome-types
```
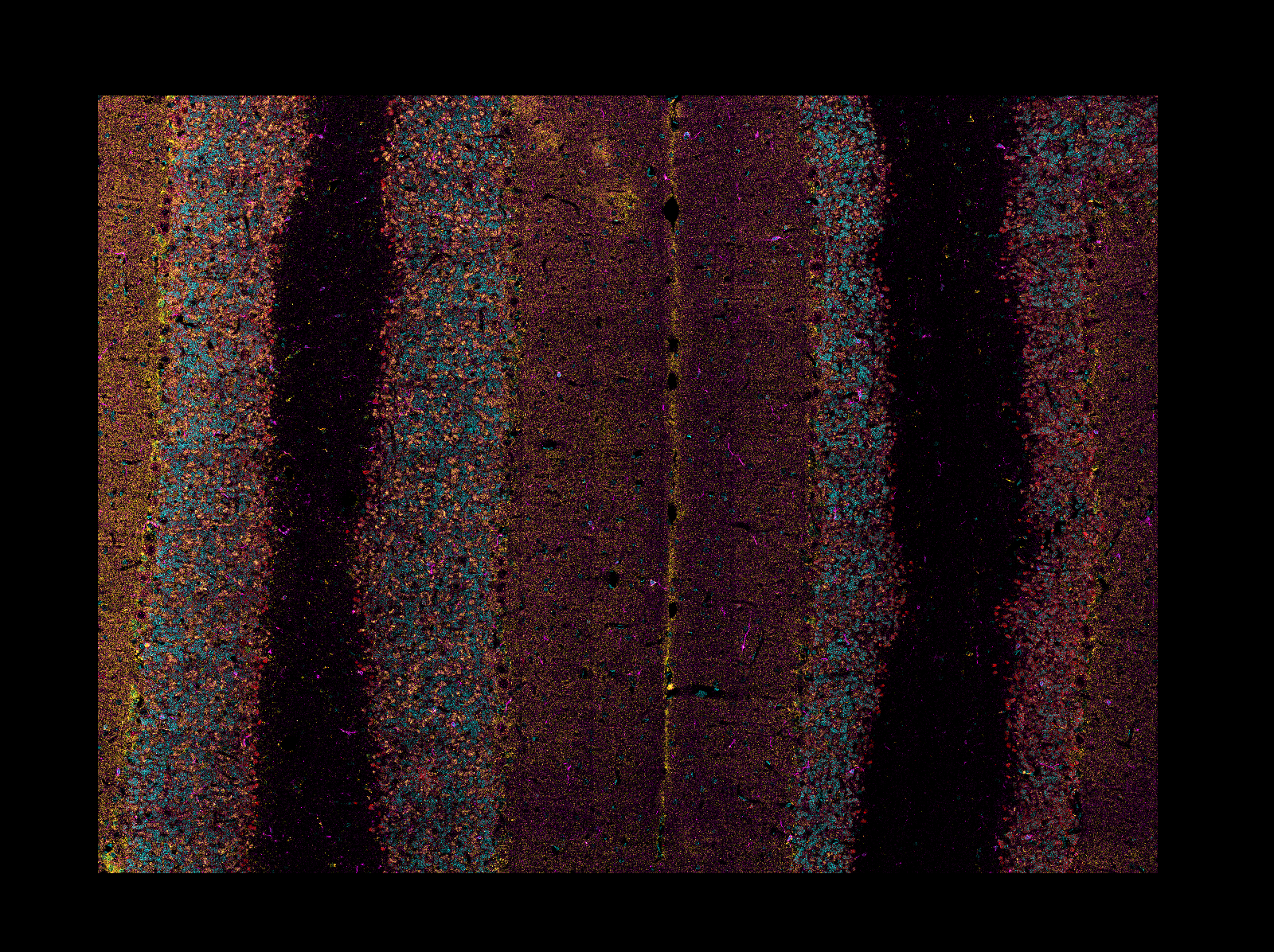
#### Mosaic Reading
When reading CZI or LIF images, if the image is a mosaic tiled image, `napari-aicsimageio`
will return the reconstructed image:
## Development
See [CONTRIBUTING.md](CONTRIBUTING.md) for information related to developing the code.
For additional file format support, contributed directly to
[AICSImageIO](https://github.com/AllenCellModeling/aicsimageio).
New file format support will become directly available in this
plugin on new `aicsimageio` releases.
## Citation
If you find `aicsimageio` and `napari-aicsimageio` useful, please cite this work as:
> Eva Maxfield Brown, Dan Toloudis, Jamie Sherman, Madison Swain-Bowden, Talley Lambert, AICSImageIO Contributors (2021). AICSImageIO: Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Pure Python [Computer software]. GitHub. https://github.com/AllenCellModeling/aicsimageio
> Eva Maxfield Brown, Talley Lambert, Peter Sobolewski, Napari-AICSImageIO Contributors (2021). Napari-AICSImageIO: Image Reading in Napari using AICSImageIO [Computer software]. GitHub. https://github.com/AllenCellModeling/napari-aicsimageio
bibtex:
```bibtex
@misc{aicsimageio,
author = {Brown, Eva Maxfield and Toloudis, Dan and Sherman, Jamie and Swain-Bowden, Madison and Lambert, Talley and {AICSImageIO Contributors}},
title = {AICSImageIO: Image Reading, Metadata Conversion, and Image Writing for Microscopy Images in Pure Python},
year = {2021},
publisher = {GitHub},
url = {https://github.com/AllenCellModeling/aicsimageio}
}
@misc{napari-aicsimageio,
author = {Brown, Eva Maxfield and Lambert, Talley and Sobolewski, Peter and {Napari-AICSImageIO Contributors}},
title = {Napari-AICSImageIO: Image Reading in Napari using AICSImageIO},
year = {2021},
publisher = {GitHub},
url = {https://github.com/AllenCellModeling/napari-aicsimageio}
}
```
_Free software: GPLv3_