https://github.com/EvilGRAHAM/flatr
Tidy and analyse contingency tables
https://github.com/EvilGRAHAM/flatr
contingency-table glm r regression tidy tidy-data
Last synced: 4 months ago
JSON representation
Tidy and analyse contingency tables
- Host: GitHub
- URL: https://github.com/EvilGRAHAM/flatr
- Owner: EvilGRAHAM
- License: other
- Created: 2017-11-03T19:05:49.000Z (about 8 years ago)
- Default Branch: master
- Last Pushed: 2018-02-21T22:17:39.000Z (almost 8 years ago)
- Last Synced: 2024-12-04T09:39:12.658Z (12 months ago)
- Topics: contingency-table, glm, r, regression, tidy, tidy-data
- Language: R
- Homepage:
- Size: 36.4 MB
- Stars: 3
- Watchers: 1
- Forks: 1
- Open Issues: 0
-
Metadata Files:
- Readme: README.Rmd
- License: LICENSE
Awesome Lists containing this project
- jimsghstars - EvilGRAHAM/flatr - Tidy and analyse contingency tables (R)
README
---
output:
md_document:
variant: markdown_github
---
```{r, echo = FALSE, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "README-"
)
library(flatr)
data("lung_cancer")
```
# flatr 
[](https://travis-ci.org/EvilGRAHAM/flatr)
## Overview
`flatr` is a package designed to make the analysis of contingency tables easier.
Contingency tables are a popular means of presenting categorical data in textbooks, as they take up very little space, while still allowing to present all the data. However, this means makes it tough to run analysis on them. `flatr` helps ease this pain by turning $i \times j \times k$ contingency tables into "tidy" data.
## Functions
* `flatten_ct()` takes an $i \times j \times k$ contingency table, and turns it into a tibble.
* `goodness_of_fit()` takes a logistic or probit regression model, and does a $\chi^{2}$ Goodness of Fit Test. The test statistic is one of:
+ 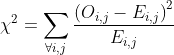
+ 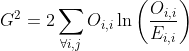
## Tidy Data
`flatr` is designed to work with the [tidyverse](https://www.tidyverse.org/) series of packages. Tidy data is data in a "long" format, where each variable has its own column.
## Usage
| Beijing | Lung | |
|--------:|------|-----|
| Smoking | Yes | No |
| Yes | 126 | 100 |
| No | 35 | 61 |
| Shanghai | Lung | |
|---------:|------|-----|
| Smoking | Yes | No |
| Yes | 908 | 688 |
| No | 497 | 807 |
| Shenyang | Lung | |
|---------:|------|-----|
| Smoking | Yes | No |
| Yes | 913 | 747 |
| No | 336 | 598 |
| Nanjing | Lung | |
|--------:|------|-----|
| Smoking | Yes | No |
| Yes | 235 | 172 |
| No | 58 | 121 |
| Harbin | Lung | |
|--------:|------|-----|
| Smoking | Yes | No |
| Yes | 402 | 308 |
| No | 121 | 215 |
| Zhengzhou | Lung | |
|----------:|------|-----|
| Smoking | Yes | No |
| Yes | 182 | 156 |
| No | 72 | 98 |
| Taiyuan | Lung | |
|--------:|------|----|
| Smoking | Yes | No |
| Yes | 60 | 99 |
| No | 11 | 43 |
| Nanchang | Lung | |
|---------:|------|----|
| Smoking | Yes | No |
| Yes | 104 | 89 |
| No | 21 | 36 |
```{r message = FALSE}
lung_tidy <- flatten_ct(lung_cancer)
lung_tidy
lung_logit <- glm(Lung ~ Smoking + City, family = binomial, data = lung_tidy)
goodness_of_fit(model = lung_logit, response = "Lung", type = "Chisq")
lung_tidy %>%
glm(
Lung ~ Smoking + City
,family = binomial(link = "probit")
,data = .
) %>%
goodness_of_fit(response = "Lung", type = "Gsq")
```