https://github.com/JhuangLab/BioInstaller
A comprehensive R package to construct interactive and reproducible biological data analysis applications based on the R platform
https://github.com/JhuangLab/BioInstaller
bioinformatics-analysis installer installer-script ngs-analysis
Last synced: 4 months ago
JSON representation
A comprehensive R package to construct interactive and reproducible biological data analysis applications based on the R platform
- Host: GitHub
- URL: https://github.com/JhuangLab/BioInstaller
- Owner: JhuangLab
- License: other
- Created: 2017-03-15T05:20:19.000Z (over 8 years ago)
- Default Branch: master
- Last Pushed: 2023-01-08T23:47:19.000Z (almost 3 years ago)
- Last Synced: 2024-10-30T21:03:03.170Z (about 1 year ago)
- Topics: bioinformatics-analysis, installer, installer-script, ngs-analysis
- Language: R
- Homepage:
- Size: 2.92 MB
- Stars: 56
- Watchers: 5
- Forks: 13
- Open Issues: 12
-
Metadata Files:
- Readme: README.md
- Changelog: ChangeLog
- License: LICENSE
Awesome Lists containing this project
- jimsghstars - JhuangLab/BioInstaller - A comprehensive R package to construct interactive and reproducible biological data analysis applications based on the R platform (R)
README
# BioInstaller 
[](https://circleci.com/gh/JhuangLab/BioInstaller/tree/master)
[](https://cran.r-project.org/package=BioInstaller)
[](https://zenodo.org/record/1343914)
[](http://www.r-pkg.org/pkg/BioInstaller)
[](https://codecov.io/github/JhuangLab/BioInstaller)
## Introduction
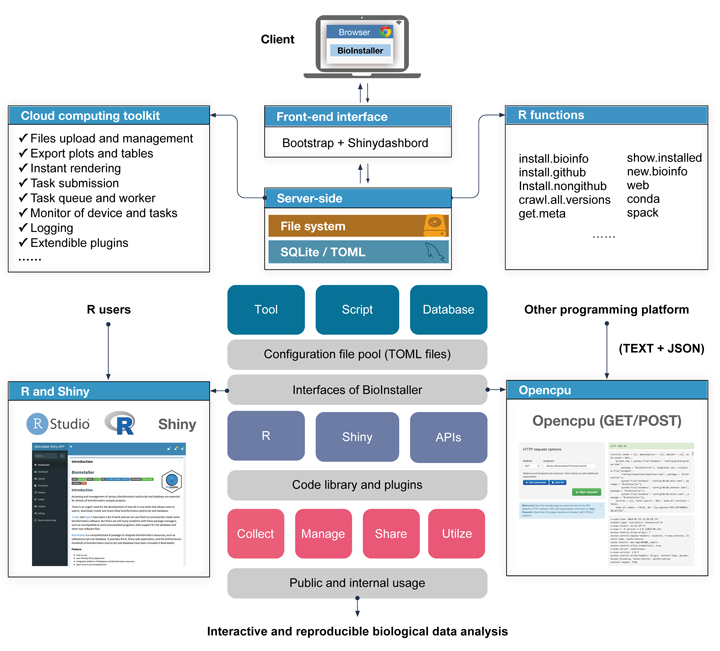
The increase in bioinformatics resources such as tools/scripts and databases poses a great challenge for users seeking to construct interactive and reproducible biological data analysis applications.
R language, as the most popular programming language for statistics, biological data analysis, and big data, has enabled diverse and free R packages (>14000) for different types of applications. However, due to the lack of high-performance and open-source cloud platforms based on R (e.g., Galaxy for Python users), it is still difficult for R users, especially those without web development skills, to construct interactive and reproducible biological data analysis applications supporting the upload and management of files, long-time computation, task submission, tracking of output files, exception handling, logging, export of plots and tables, and extendible plugin systems.
The collection, management, and share of various bioinformatics tools/scripts and databases are also essential for almost all bioinformatics analysis projects.
Here, we established a new platform to construct interactive and reproducible biological data analysis applications based on R language. This platform contains diverse user interfaces, including the R functions and R Shiny application, REST APIs, and support for collecting, managing, sharing, and utilizing massive bioinformatics tools/scripts and databases.
**Feature**:
- Easy-to-use
- User-friendly Shiny application
- Integrative platform of Databases and bioinformatics resources
- Open source and completely free
- One-click to download and install bioinformatics resources (via R, Shiny or Opencpu REST APIs)
- More attention for those software and database resource that have not been
by other tools
- Logging
- System monitor
- Task submitting system
- Parallel tasks
**Field**
- Quality Control
- Alignment And Assembly
- Alternative Splicing
- ChIP-seq analysis
- Gene Expression Data Analysis
- Variant Detection
- Variant Annotation
- Virus Related
- Statistical and Visualization
- Noncoding RNA Related Database
- Cancer Genomics Database
- Regulator Related Database
- eQTL Related Database
- Clinical Annotation
- Drugs Database
- Proteomic Database
- Software Dependence Database
- ......
**Note:** We are developing [bget](https://github.com/openbiox/bget) and [bioshiny](https://github.com/openbiox/bioshiny) projects independently for simplify the functions of download and shiny.
- [bget](https://github.com/openbiox/bget) is an golang-based command-line tool that do not need to install any R packages.
- [bioshiny](https://github.com/openbiox/bioshiny) is the core shiny application of previous BioInstaller package.
## Installation
### CRAN
``` r
#You can install this package directly from CRAN by running (from within R):
install.packages('BioInstaller')
```
### Github
``` bash
# install.packages("devtools")
devtools::install_github("JhuangLab/BioInstaller")
```
## Shiny application
**Note**, the Shiny application of BioInstaller was migrated to [bioshiny](https://github.com/openbiox/bioshiny) project. All shiny files in this package have been removed for reducing package size.
In the new project, we are developing more free plugins of bioshiny for various bioinformatics data analysis.
```bash
echo 'export BIO_SOFTWARES_DB_ACTIVE="~/.bioshiny/info.yaml" >> ~/.bashrc'
echo 'export BIOSHINY_CONFIG="~/.bioshiny/shiny.config.yaml" >> ~/.bashrc'
. ~/.bashrc
# Start the standalone Shiny application
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_deps_r
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_start
chmod a+x bioshiny_deps_r
chmod a+x bioshiny_start
./bioshiny_deps_r
# Start Shiny application workers
Rscript -e "bioshiny::set_shiny_workers(1)"
./bioshiny_start
# or use yarn
yarn global add bioshiny
bioshiny_deps_r
Rscript -e "bioshiny::set_shiny_workers(1)"
bioshiny_start
```
[spack](https://spack.io/) and [miniconda](https://docs.conda.io/en/latest/miniconda.html) are required for extra functions.
## Contributed Resources
- [GitHub
resource](https://github.com/JhuangLab/BioInstaller/blob/master/inst/extdata/config/github/github.toml)
- GitHub resource [meta
information](https://github.com/JhuangLab/BioInstaller/blob/master/inst/extdata/config/github/github_meta.toml)
- [Non GitHub
resource](https://github.com/JhuangLab/BioInstaller/blob/master/inst/extdata/config/nongithub/nongithub.toml)
- Non Github resource [meta
infrmation](https://github.com/JhuangLab/BioInstaller/blob/master/inst/extdata/config/nongithub/nongithub_meta.toml)
- [Database](https://github.com/JhuangLab/BioInstaller/tree/master/inst/extdata/config/db)
- [Web
Service](https://github.com/JhuangLab/BioInstaller/blob/master/inst/extdata/config/web/web_meta.toml)
- [Docker](https://github.com/JhuangLab/BioInstaller/blob/master/inst/extdata/config/docker/docker.toml)
## Support Summary
**Quality Control:**
- FastQC, PRINSEQ, SolexaQA, FASTX-Toolkit ...
**Alignment and Assembly:**
- BWA, STAR, TMAP, Bowtie, Bowtie2, tophat2, hisat2, GMAP-GSNAP,
ABySS, SSAHA2, Velvet, Edean, Trinity, oases, RUM, MapSplice2,
NovoAlign ...
**Variant Detection:**
- GATK, Mutect, VarScan2, FreeBayes, LoFreq, TVC, SomaticSniper,
Pindel, Delly, BreakDancer, FusionCatcher, Genome STRiP, CNVnator,
CNVkit, SpeedSeq ...
**Variant Annotation:**
- ANNOVAR, SnpEff, VEP, oncotator ...
**Utils:**
- htslib, samtools, bcftools, bedtools, bamtools, vcftools, sratools,
picard, HTSeq, seqtk, UCSC Utils(blat, liftOver), bamUtil, jvarkit,
bcl2fastq2, fastq\_tools ...
**Genome:**
- hisat2\_reffa, ucsc\_reffa, ensemble\_reffa ...
**Others:**
- sparsehash, SQLite, pigz, lzo, lzop, bzip2, zlib, armadillo, pxz,
ROOT, curl, xz, pcre, R, gatk\_bundle, ImageJ, igraph ...
**Databases:**
- ANNOVAR, blast, CSCD, GATK\_Bundle, biosystems, civic, denovo\_db,
dgidb, diseaseenhancer, drugbank, ecodrug, expression\_atlas,
funcoup, gtex, hpo, inbiomap, interpro, medreaders, mndr, msdd,
omim, pancanqtl, proteinatlas, remap2, rsnp3, seecancer,
srnanalyzer, superdrug2, tumorfusions, varcards ...
## Docker
You can use the BioInstaller in Docker since v0.3.0. Shiny application was supported since v0.3.5.
``` bash
docker pull bioinstaller/bioinstaller
docker run -it -p 80:80 -p 8004:8004 -v /tmp/download:/tmp/download bioinstaller/bioinstaller
```
Service list:
- localhost/ocpu/ Opencpu service
- localhost/shiny/BioInstaller Shiny service
- localhost/rstudio/ Rstudio server (opencpu/opencpu)
## Citation
- Li J, Cui B, Dai Y, et al. BioInstaller: a comprehensive R package to construct interactive and reproducible biological data analysis applications based on the R platform[J]. PeerJ, 2018, 6:e5853.
## How to contribute?
Please fork the [GitHub BioInstaller
repository](https://github.com/JhuangLab/BioInstaller), modify it, and
submit a pull request to us. Especialy, the files list in `contributed
section` should be modified when you see a tool or database that not be
included in the other software warehouse.
## Maintainer
[Jianfeng Li](https://github.com/Miachol)
## License
R package:
[MIT](https://en.wikipedia.org/wiki/MIT_License)
Related Other Resources
Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License