https://github.com/Merck/r2rtf
Easily Create Production-Ready Rich Text Format (RTF) Table and Figure
https://github.com/Merck/r2rtf
Last synced: 4 months ago
JSON representation
Easily Create Production-Ready Rich Text Format (RTF) Table and Figure
- Host: GitHub
- URL: https://github.com/Merck/r2rtf
- Owner: Merck
- License: gpl-3.0
- Created: 2020-02-19T22:10:51.000Z (almost 6 years ago)
- Default Branch: master
- Last Pushed: 2024-09-30T21:18:17.000Z (over 1 year ago)
- Last Synced: 2025-01-13T01:02:53.354Z (about 1 year ago)
- Language: R
- Homepage: https://merck.github.io/r2rtf
- Size: 11.3 MB
- Stars: 78
- Watchers: 11
- Forks: 20
- Open Issues: 6
-
Metadata Files:
- Readme: README.Rmd
- Changelog: NEWS.md
- License: LICENSE
- Citation: CITATION.cff
Awesome Lists containing this project
- top-pharma50 - **Merck/r2rtf** - Ready Rich Text Format (RTF) Table and Figure<br><img src='https://github.com/HubTou/topgh/blob/main/icons/gstars.png'> 76 <img src='https://github.com/HubTou/topgh/blob/main/icons/forks.png'> 19 <img src='https://github.com/HubTou/topgh/blob/main/icons/code.png'> R <img src='https://github.com/HubTou/topgh/blob/main/icons/license.png'> GNU General Public License v3.0 <img src='https://github.com/HubTou/topgh/blob/main/icons/last.png'> 2024-06-03 01:28:44 | (Ranked by starred repositories)
README
---
output: github_document
---
# r2rtf 
[](https://CRAN.R-project.org/package=r2rtf)
[](https://app.codecov.io/gh/Merck/r2rtf)
[](https://cran.r-project.org/package=r2rtf)
[](https://github.com/Merck/r2rtf/actions/workflows/R-CMD-check.yaml)
[](https://cran.r-project.org/package=r2rtf)
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
```
```{r, echo = FALSE}
pkgname <- "r2rtf"
```
## Overview
r2rtf is an R package to create production-ready tables and figures in RTF format.
The package is designed with these principles:
- Provide simple "verb" functions that correspond to each component of a table,
to help you translate data frame(s) to a table in RTF file.
- Functions are chainable with pipes (`%>%`).
- Only focus on **table format**.
- Data manipulation and analysis should be handled by other R packages, for example, tidyverse.
- Minimize package dependency.
The [R for clinical study reports and submission](https://r4csr.org/) book
provides tutorials by using real world examples.
## Installation
You can install the package via CRAN:
```r
install.packages("r2rtf")
```
Or, install from GitHub:
```r
remotes::install_github("Merck/r2rtf")
```
## Highlighted features
The R package r2rtf provides flexibility to enable features below:
- Create highly customized RTF tables and figures ready for production.
- Simple to use parameters and data structure.
- Customized column header: split by `"|"`.
- Three required parameters for the output tables (data, filename, column relative width).
- Flexible and detail control of table structure.
- Format control in cell, row, column and table level for:
- Border Type: single, double, dash, dot, etc.
- Alignment: left, right, center, decimal.
- Column width.
- Text appearance: **bold**, *italics*, ~~strikethrough~~, underline and any combinations.
- Font size.
- Text and border color (`r length(colors())` different colors named in `color()` function).
- Special characters: any character in UTF-8 encoding (e.g., Greek, Symbol, Chinese, Japanese, Korean).
- Append several tables into one file.
- Pagination.
- Built-in raw data for validation.
## Simple example
```{r, eval = FALSE}
library(dplyr)
library(r2rtf)
head(iris) %>%
rtf_body() %>% # Step 1 Add attributes
rtf_encode() %>% # Step 2 Convert attributes to RTF encode
write_rtf(file = "ex-tbl.rtf") # Step 3 Write to a .rtf file
```
```{r, include=FALSE}
library(r2rtf)
try(
{
pdftools::pdf_convert("vignettes/pdf/ex-tbl.pdf", dpi = 200, filenames = "ex-tbl.png")
pdftools::pdf_convert("vignettes/pdf/efficacy_example.pdf", dpi = 200, filenames = "efficacy_example.png")
pdftools::pdf_convert("vignettes/pdf/ae_example.pdf", dpi = 200, filenames = "ae_example.png")
files <- c("ex-tbl.png", "efficacy_example.png", "ae_example.png")
file.copy(from = files, to = file.path("vignettes/fig", files), overwrite = TRUE)
file.remove(files)
},
silent = TRUE
)
```
Click here to see the output
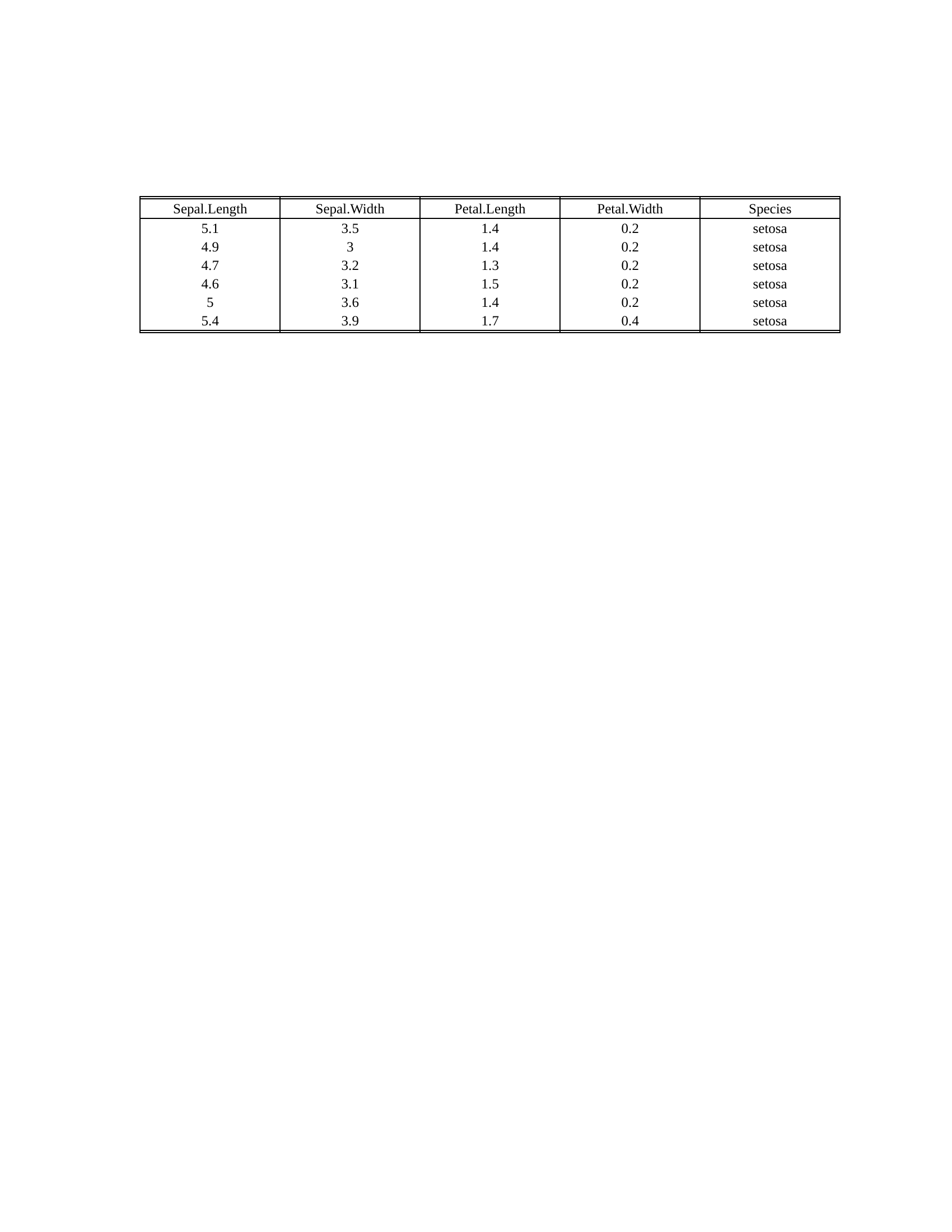
- [More Examples](https://merck.github.io/r2rtf/articles/index.html)
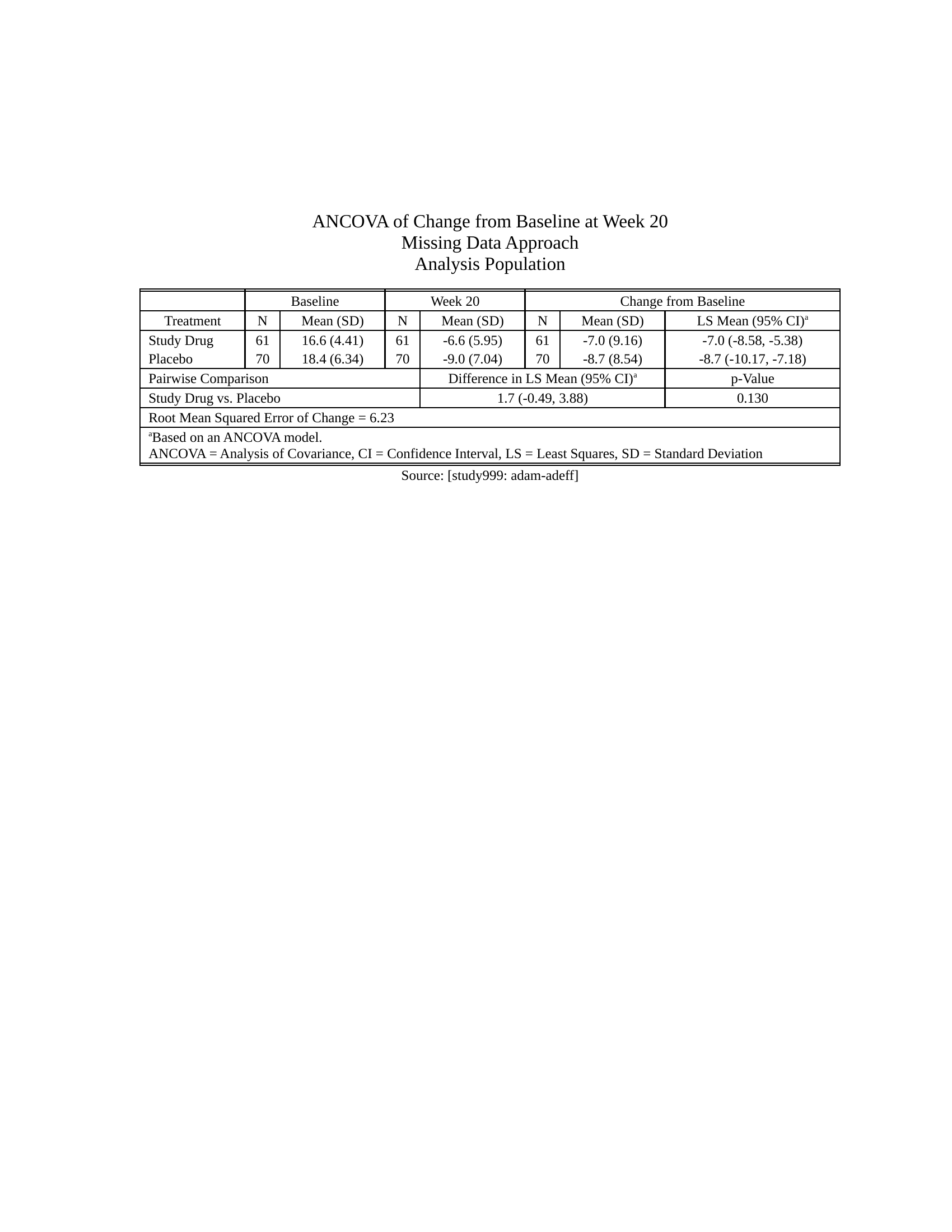
## Example efficacy table
- [Source code](https://merck.github.io/r2rtf/articles/example-efficacy.html)
Click here to see the output
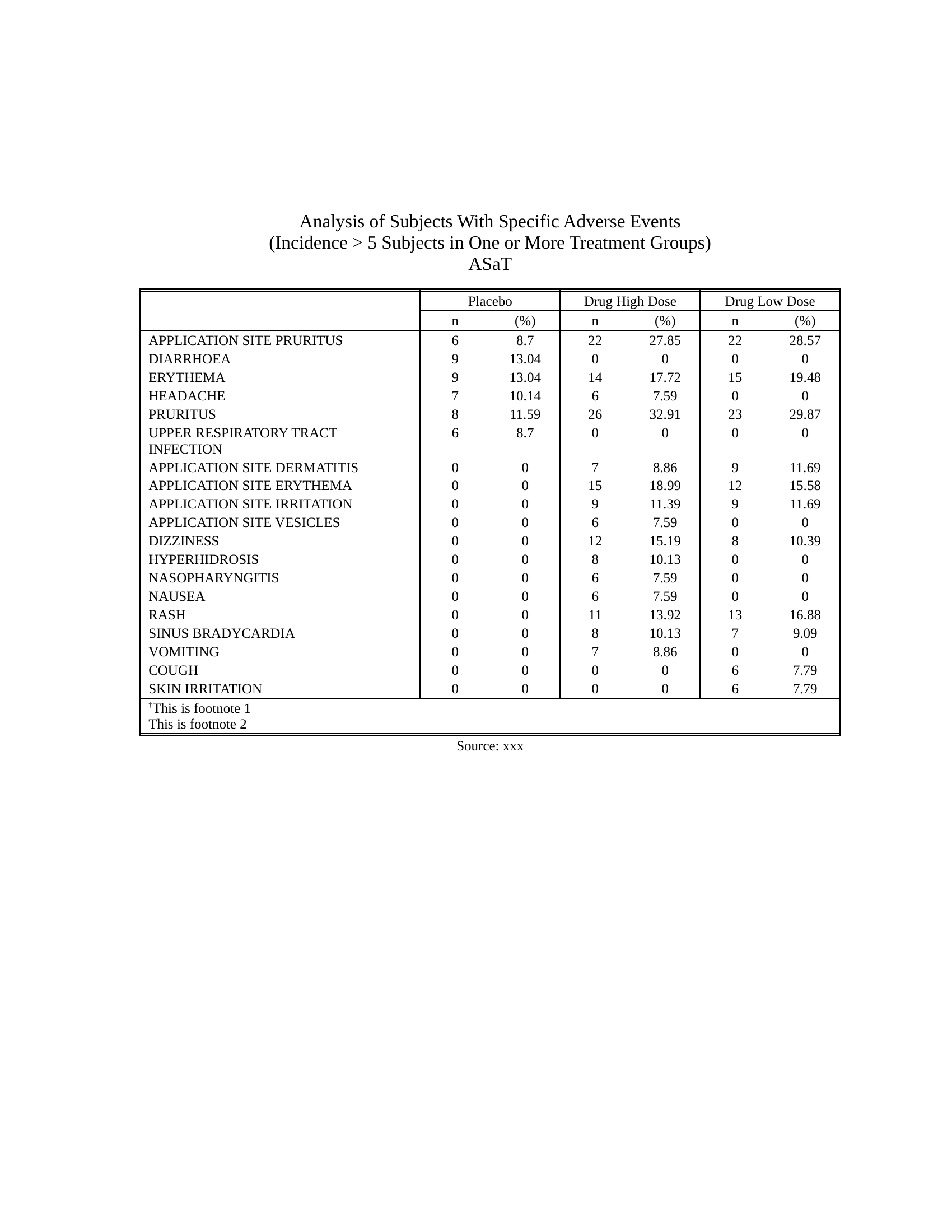
## Example safety table
- [Source code](https://merck.github.io/r2rtf/articles/example-ae-summary.html)
Click here to see the output
```{r, include=FALSE, eval = FALSE}
# Transfer all RTF files to PDF files
files <- list.files("vignettes/rtf", pattern = "*.rtf", full.names = TRUE)
r2rtf:::rtf_convert_format(
input = files,
format = "pdf",
output_dir = "vignettes/pdf"
)
r2rtf:::rtf_convert_format(
input = files,
format = "docx",
output_dir = "vignettes/docx"
)
r2rtf:::rtf_convert_format(
input = files,
format = "html",
output_dir = "vignettes/html"
)
files <- list.files("vignettes/rtf", pattern = "*\\.rtf")
files_pdf <- list.files("vignettes/pdf", pattern = "*\\.pdf")
files_docx <- list.files("vignettes/docx", pattern = "*\\.docx")
files_html <- list.files("vignettes/html", pattern = "*\\.html")
# Compare RTF and PDF folder filenames.
waldo::compare(
tools::file_path_sans_ext(files),
tools::file_path_sans_ext(files_pdf)
)
waldo::compare(
tools::file_path_sans_ext(files),
tools::file_path_sans_ext(files_docx)
)
waldo::compare(
tools::file_path_sans_ext(files),
tools::file_path_sans_ext(files_html)
)
```
## Citation
If you use this software, please cite it as below.
> Wang, S., Ye, S., Anderson, K., & Zhang, Y. (2020).
> r2rtf---an R Package to Produce Rich Text Format (RTF) Tables and Figures.
> _PharmaSUG_. https://pharmasug.org/proceedings/2020/DV/PharmaSUG-2020-DV-198.pdf
A BibTeX entry for LaTeX users is
```bibtex
@inproceedings{wang2020r2rtf,
title = {{r2rtf}---an {R} Package to Produce {Rich Text Format} ({RTF}) Tables and Figures},
author = {Wang, Siruo and Ye, Simiao and Anderson, Keaven M and Zhang, Yilong},
booktitle = {PharmaSUG},
year = {2020},
url = {https://pharmasug.org/proceedings/2020/DV/PharmaSUG-2020-DV-198.pdf}
}
```