Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/MontaEllis/Pytorch-Medical-Segmentation
This repository is an unoffical PyTorch implementation of Medical segmentation in 2D and 3D.
https://github.com/MontaEllis/Pytorch-Medical-Segmentation
medical-image-processing pytorch segmentation torchio
Last synced: 2 months ago
JSON representation
This repository is an unoffical PyTorch implementation of Medical segmentation in 2D and 3D.
- Host: GitHub
- URL: https://github.com/MontaEllis/Pytorch-Medical-Segmentation
- Owner: MontaEllis
- License: mit
- Created: 2021-01-08T04:25:42.000Z (about 4 years ago)
- Default Branch: master
- Last Pushed: 2024-02-29T11:05:38.000Z (11 months ago)
- Last Synced: 2024-08-08T23:21:36.314Z (6 months ago)
- Topics: medical-image-processing, pytorch, segmentation, torchio
- Language: Python
- Homepage:
- Size: 85 KB
- Stars: 838
- Watchers: 11
- Forks: 196
- Open Issues: 9
-
Metadata Files:
- Readme: README-zh.md
- License: LICENSE
Awesome Lists containing this project
README
# Pytorch Medical Segmentation
英文版请戳:这里!
## Notes
我们计划在最近对代码进行一次大更新,如果您有任何建议,请随意给[我]([email protected])发邮件或者在issue里提。
## 最近的更新
* 2021.1.8 训练和测试代码已经发布
* 2021.2.6 修复计算dice的一个bug,感谢[Shanshan Li](https://github.com/ssli23)的帮助~
* 2021.2.24 发布一个视频教程(https://www.bilibili.com/video/BV1gp4y1H7kq/)。
* 2021.5.16 修复Unet3D实现的错误。
* 2021.5.16 评估代码公布。
* 2021.6.24 所有参数都能在hparam.py中调整。
* 2021.7.7 现在你可以参考医学分类程序: [Pytorch-Medical-Classification](https://github.com/MontaEllis/Pytorch-Medical-Classification)
* 2022.5.15 现在你可以参考半监督医学分割程序: [SSL-For-Medical-Segmentation](https://github.com/MontaEllis/SSL-For-Medical-Segmentation)
* 2022.5.17 我们更新了训练和测试代码,修复了一些小bug。
## 环境要求
* pytorch1.7
* torchio<=0.18.20
* python>=3.6
## 通知
* 您可以修改**hparam.py**文件来确定是2D分割还是3D分割以及是否可以进行多分类。
* 我们几乎提供了所有的2D和3D分割的算法。
* 本项目兼容几乎所有的医学数据格式(例如 nii.gz, nii, mhd, nrrd, ...),修改**hparam.py**的**fold_arch**即可。**我希望您能在使用前把source和label图片都转成相同的类型,其中,label用1标志,不是255。**
* 如果您想进行**多分类**分割,请自行修改对应代码。我不能确定您的具体分类数。
* 不论是2D或是3D,本项目均采用**patch**的方式。故图片大小不必严格保持一致。在2D中,您应该把patch设置的足够大。
## 准备您的数据
### 例1
如果您的source文件夹如下排列 :
```
source_dataset
├── source_1.mhd
├── source_1.zraw
├── source_2.mhd
├── source_2.zraw
├── source_3.mhd
├── source_3.zraw
├── source_4.mhd
├── source_4.zraw
└── ...
```
同时您的label文件夹如下排列 :
```
label_dataset
├── label_1.mhd
├── label_1.zraw
├── label_2.mhd
├── label_2.zraw
├── label_3.mhd
├── label_3.zraw
├── label_4.mhd
├── label_4.zraw
└── ...
```
您应该修改 **fold_arch** 为 **\*.mhd**, **source_train_dir** 为 **source_dataset** 并修改 **label_train_dir** 为 **label_dataset** in **hparam.py**
### Example2
如果您的source文件夹如下排列 :
```
source_dataset
├── 1
├── source_1.mhd
├── source_1.zraw
├── 2
├── source_2.mhd
├── source_2.zraw
├── 3
├── source_3.mhd
├── source_3.zraw
├── 4
├── source_4.mhd
├── source_4.zraw
└── ...
```
同时您的label文件夹如下排列 :
```
label_dataset
├── 1
├── label_1.mhd
├── label_1.zraw
├── 2
├── label_2.mhd
├── label_2.zraw
├── 3
├── label_3.mhd
├── label_3.zraw
├── 4
├── label_4.mhd
├── label_4.zraw
└── ...
```
您应该修改 **fold_arch** 为 **\*/\*.mhd**, **source_train_dir** 为 **source_dataset** 并修改 **label_train_dir** 为 **label_dataset** in **hparam.py**
## 训练
* 不使用预训练模型
```
set hparam.train_or_test to 'train'
python main.py
```
* 使用预训练模型
```
set hparam.train_or_test to 'train'
python main.py -k True
```
## Inference
* 测试
```
set hparam.train_or_test to 'test'
python main.py
```
## 实例
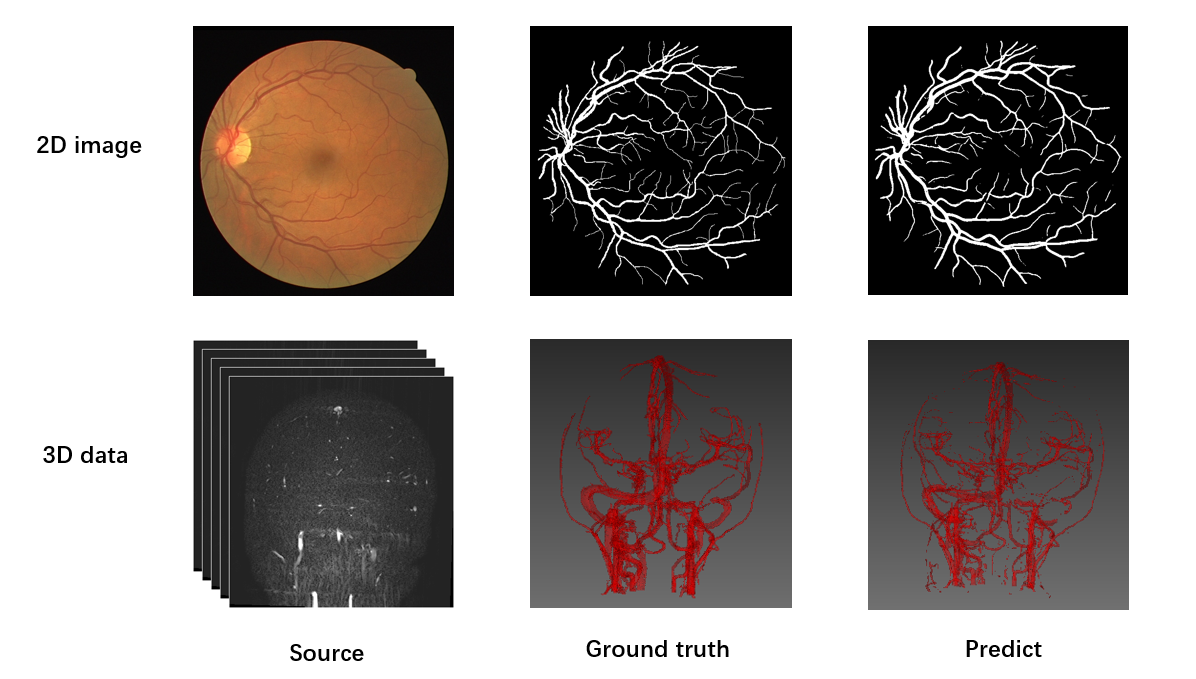
## 教程
* https://www.bilibili.com/video/BV1gp4y1H7kq/
## Done
### Network
* 2D
- [x] unet
- [x] unet++
- [x] miniseg
- [x] segnet
- [x] pspnet
- [x] highresnet(copy from https://github.com/fepegar/highresnet, Thank you to [fepegar](https://github.com/fepegar) for your generosity!)
- [x] deeplab
- [x] fcn
* 3D
- [x] unet3d
- [x] residual-unet3d
- [x] densevoxelnet3d
- [x] fcn3d
- [x] vnet3d
- [x] highresnert(copy from https://github.com/fepegar/highresnet, Thank you to [fepegar](https://github.com/fepegar) for your generosity!)
- [x] densenet3d
- [x] unetr (copy from https://github.com/tamasino52/UNETR)
### Metric
- [x] metrics.py 来评估您的结果
## TODO
- [ ] dataset
- [ ] benchmark
- [ ] nnunet
## By The Way
这个项目并不完美,还存在很多问题。如果您正在使用这个项目,并想给作者一些反馈,您可以给[我]([email protected])发邮件。
## 致谢
这个项目是一个非官方PyTorch实现的3D和2D医学分割,高度依赖于[MedicalZooPytorch](https://github.com/black0017/MedicalZooPytorch)和[torchio](https://github.com/fepegar/torchio)。感谢上述项目。本项目在[Prof. Ruoxiu Xiao](http://enscce.ustb.edu.cn/Teach/TeacherList/2020-10-16/114.html), [Prof. Shuang Song]([email protected]) 和 [Dr. Cheng Chen]([email protected])的指导下完成。感谢[Youming Zhang]([email protected]), [Daiheng Gao](https://github.com/tomguluson92), [Jie Zhang]([email protected]), [Xing Tao]([email protected]), [Weili Jiang]([email protected])和[Shanshan Li](https://github.com/ssli23) 对我的帮助。