Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/SciML/SciMLTutorials.jl
Tutorials for doing scientific machine learning (SciML) and high-performance differential equation solving with open source software.
https://github.com/SciML/SciMLTutorials.jl
dae dde differential-equations differentialequations hacktoberfest julia neural-differential-equations neural-ode ode ordinary-differential-equations partial-differential-equations pde python r scientific-machine-learning sciml sde stochastic-differential-equations
Last synced: about 2 months ago
JSON representation
Tutorials for doing scientific machine learning (SciML) and high-performance differential equation solving with open source software.
- Host: GitHub
- URL: https://github.com/SciML/SciMLTutorials.jl
- Owner: SciML
- License: other
- Created: 2016-10-31T04:04:42.000Z (almost 8 years ago)
- Default Branch: master
- Last Pushed: 2023-09-04T18:42:26.000Z (about 1 year ago)
- Last Synced: 2024-04-14T01:46:57.799Z (5 months ago)
- Topics: dae, dde, differential-equations, differentialequations, hacktoberfest, julia, neural-differential-equations, neural-ode, ode, ordinary-differential-equations, partial-differential-equations, pde, python, r, scientific-machine-learning, sciml, sde, stochastic-differential-equations
- Language: CSS
- Homepage: https://tutorials.sciml.ai
- Size: 217 MB
- Stars: 706
- Watchers: 25
- Forks: 128
- Open Issues: 14
-
Metadata Files:
- Readme: README.md
- License: LICENSE.md
- Citation: CITATION.bib
Awesome Lists containing this project
README
# SciMLTutorials.jl: Tutorials for Scientific Machine Learning and Differential Equations
[](https://julialang.zulipchat.com/#narrow/stream/279055-sciml-bridged)
[](http://tutorials.sciml.ai/stable/)
[](https://docs.sciml.ai/dev/highlevels/learning_resources/#SciMLTutorials)
[](https://buildkite.com/julialang/scimltutorials-dot-jl)
[](https://github.com/SciML/ColPrac)
[](https://github.com/SciML/SciMLStyle)
SciMLTutorials.jl holds PDFs, webpages, and interactive Jupyter notebooks
showing how to utilize the software in the [SciML Scientific Machine Learning ecosystem](https://sciml.ai/).
This set of tutorials was made to complement the [documentation](https://sciml.ai/documentation/)
and the [devdocs](http://devdocs.sciml.ai/latest/)
by providing practical examples of the concepts. For more details, please
consult the docs.
#### Note: this library has been deprecated and its tutorials have been moved to the repos of the respective packages. It may be revived in the future if there is a need for longer-form tutorials!
## Results
To view the SciML Tutorials, go to [tutorials.sciml.ai](https://tutorials.sciml.ai/stable/). By default, this
will lead to the latest tagged version of the tutorials. To see the in-development version of the tutorials, go to
[https://tutorials.sciml.ai/dev/](https://tutorials.sciml.ai/dev/).
Static outputs in pdf, markdown, and html reside in [SciMLTutorialsOutput](https://github.com/SciML/SciMLTutorialsOutput).
## Video Tutorial
[](https://youtu.be/KPEqYtEd-zY)
## Interactive Notebooks
To generate the interactive notebooks, first install the SciMLTutorials, instantiate the
environment, and then run `SciMLTutorials.open_notebooks()`. This looks as follows:
```julia
]add SciMLTutorials#master
]activate SciMLTutorials
]instantiate
using SciMLTutorials
SciMLTutorials.open_notebooks()
```
The tutorials will be generated at your `pwd()` in a folder called `generated_notebooks`.
Note that when running the tutorials, the packages are not automatically added. Thus you
will need to add the packages manually or use the internal Project/Manifest tomls to
instantiate the correct packages. This can be done by activating the folder of the tutorials.
For example,
```julia
using Pkg
Pkg.activate(joinpath(pkgdir(SciMLTutorials),"tutorials","models"))
Pkg.instantiate()
```
will add all of the packages required to run any tutorial in the `models` folder.
## Contributing
All of the files are generated from the Weave.jl files in the `tutorials` folder. The generation process runs automatically,
and thus one does not necessarily need to test the Weave process locally. Instead, simply open a PR that adds/updates a
file in the "tutorials" folder and the PR will generate the tutorial on demand. Its artifacts can then be inspected in the
Buildkite as described below before merging. Note that it will use the Project.toml and Manifest.toml of the subfolder, so
any changes to dependencies requires that those are updated.
### Reporting Bugs and Issues
Report any bugs or issues at [the SciMLTutorials repository](https://github.com/SciML/SciMLTutorials.jl/issues).
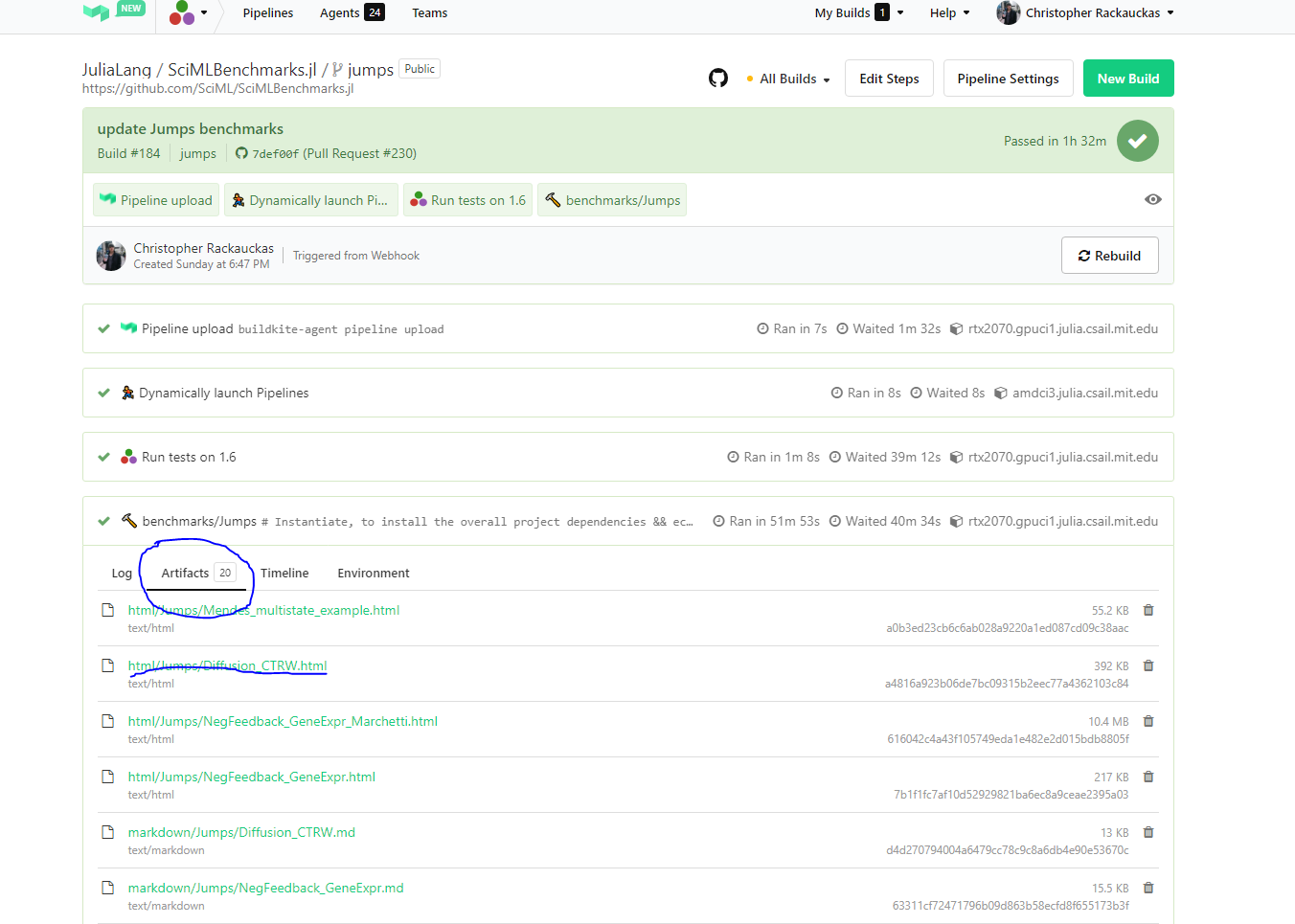
### Inspecting Tutorial Results
To see tutorial results before merging, click into the BuildKite, click onto
Artifacts, and then investigate the trained results.
### Manually Generating Files
To run the generation process, do for example:
```julia
]activate SciMLTutorials # Get all of the packages
using SciMLTutorials
SciMLTutorials.weave_file(joinpath(pkgdir(SciMLTutorials),"tutorials","models"),"01-classical_physics.jmd")
```
To generate all of the files in a folder, for example, run:
```julia
SciMLTutorials.weave_folder(joinpath(pkgdir(SciMLTutorials),"tutorials","models"))
```
To generate all of the notebooks, do:
```julia
SciMLTutorials.weave_all()
```
Each of the tuturials displays the computer characteristics at the bottom of
the benchmark.