https://github.com/adosar/aidsorb
Python package for deep learning on molecular point clouds.
https://github.com/adosar/aidsorb
3d-deep-learning 3d-point-clouds deep-learning geometric-deep-learning machine-learning material-informatics metal-organic-frameworks point-clouds pytorch pytorch-lightning
Last synced: 8 months ago
JSON representation
Python package for deep learning on molecular point clouds.
- Host: GitHub
- URL: https://github.com/adosar/aidsorb
- Owner: adosar
- License: gpl-3.0
- Created: 2024-02-22T13:22:05.000Z (almost 2 years ago)
- Default Branch: master
- Last Pushed: 2025-03-20T22:15:15.000Z (10 months ago)
- Last Synced: 2025-04-12T13:37:28.279Z (9 months ago)
- Topics: 3d-deep-learning, 3d-point-clouds, deep-learning, geometric-deep-learning, machine-learning, material-informatics, metal-organic-frameworks, point-clouds, pytorch, pytorch-lightning
- Language: Python
- Homepage: https://aidsorb.readthedocs.io/en/stable/
- Size: 16.7 MB
- Stars: 2
- Watchers: 1
- Forks: 1
- Open Issues: 8
-
Metadata Files:
- Readme: README.md
- License: LICENSE
- Citation: CITATION.cff
Awesome Lists containing this project
README







[](https://aidsorb.readthedocs.io/en/stable/)
[](https://pypi.org/project/aidsorb/)
[](https://aidsorb-online.streamlit.app)
**AIdsorb** is a Python package for **deep learning on molecular point clouds**.
This package aims to provide a **simple, easy-to-use and reproduce** interface for:
- 📥 **Creating molecular point clouds**
- 🤖 **Training DL algorithms on molecular point clouds**
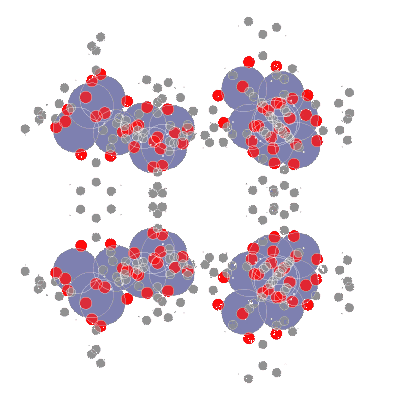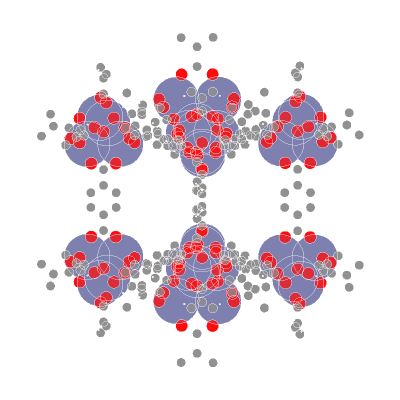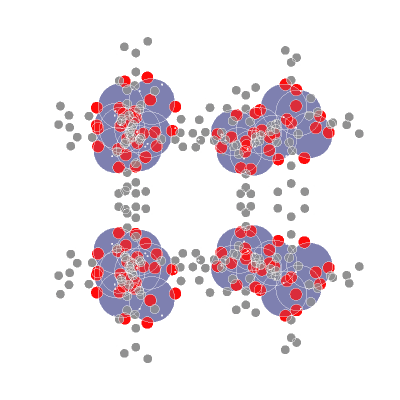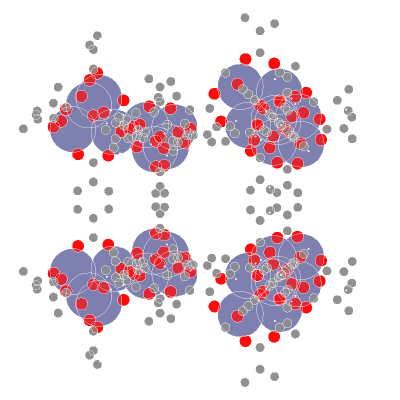
## ⚙️ Installation
> [!IMPORTANT]
> It is strongly recommended to **perform the installation inside a virtual environment**.
Assuming an activated virtual environment:
```bash
pip install aidsorb
```
## 🚀 Usage
> [!NOTE]
> Refer to the 📚 [Documentation](https://aidsorb.readthedocs.io/en/stable/) for more information.
Here is a summary of what you can do from the command line:
1. Visualize a molecular point cloud:
```bash
aidsorb visualize path/to/structure
```
2. Create and prepare point clouds:
```bash
aidsorb create path/to/structures path/to/pcd_data # Create and store point clouds
aidsorb prepare path/to/pcd_data # Split point clouds to train, valdation and test
```
3. Train and test a model:
```bash
aidsorb-lit fit --config=path/to/config.yaml
aidsorb-lit test --config=path/to/config.yaml --ckpt_path=path/to/ckpt
```
## 💡 Contributing
🙌 We welcome contributions from the community to help improve and expand this
project!
You can start by 🛠️ [opening an issue](https://github.com/adosar/aidsorb/issues) for:
* 🐛 Reporting bugs
* 🌟 Suggesting new features
* 📚 Improving documentation
* 🎨 Adding your example to the [Gallery](https://aidsorb.readthedocs.io/en/stable/auto_examples/index.html)
We appreciate your efforts to submit well-documented 🔃 [pull
requests](https://github.com/adosar/aidsorb/pulls) and participate in
discussions.
💪 Together, we can make this project even better!
## 📑 Citing
* **To cite the software**, please refer to the [citation file](./CITATION.cff) or click the citation button.
* **To cite the paper**, please use the following BibTeX entry:
Show BibTex entry
```bibtex
@article{Sarikas2024,
title = {Gas adsorption meets geometric deep learning: points, set and match},
volume = {14},
ISSN = {2045-2322},
url = {http://dx.doi.org/10.1038/s41598-024-76319-8},
DOI = {10.1038/s41598-024-76319-8},
number = {1},
journal = {Scientific Reports},
publisher = {Springer Science and Business Media LLC},
author = {Sarikas, Antonios P. and Gkagkas, Konstantinos and Froudakis, George E.},
year = {2024},
month = nov
}
```
## ⚖️ License
**AIdosrb** is released under the [GNU General Public License v3.0 only](https://spdx.org/licenses/GPL-3.0-only.html).