https://github.com/andim/paper-tcellexp
Source code accompanying the paper "Regulation of T cell expansion by antigen presentation dynamics "
https://github.com/andim/paper-tcellexp
biophysics openscience
Last synced: about 1 month ago
JSON representation
Source code accompanying the paper "Regulation of T cell expansion by antigen presentation dynamics "
- Host: GitHub
- URL: https://github.com/andim/paper-tcellexp
- Owner: andim
- License: mit
- Created: 2019-05-17T21:17:49.000Z (over 6 years ago)
- Default Branch: master
- Last Pushed: 2019-05-17T21:33:48.000Z (over 6 years ago)
- Last Synced: 2025-09-09T06:41:53.950Z (5 months ago)
- Topics: biophysics, openscience
- Language: Python
- Size: 916 KB
- Stars: 0
- Watchers: 2
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Regulation of T cell expansion by antigen presentation dynamics
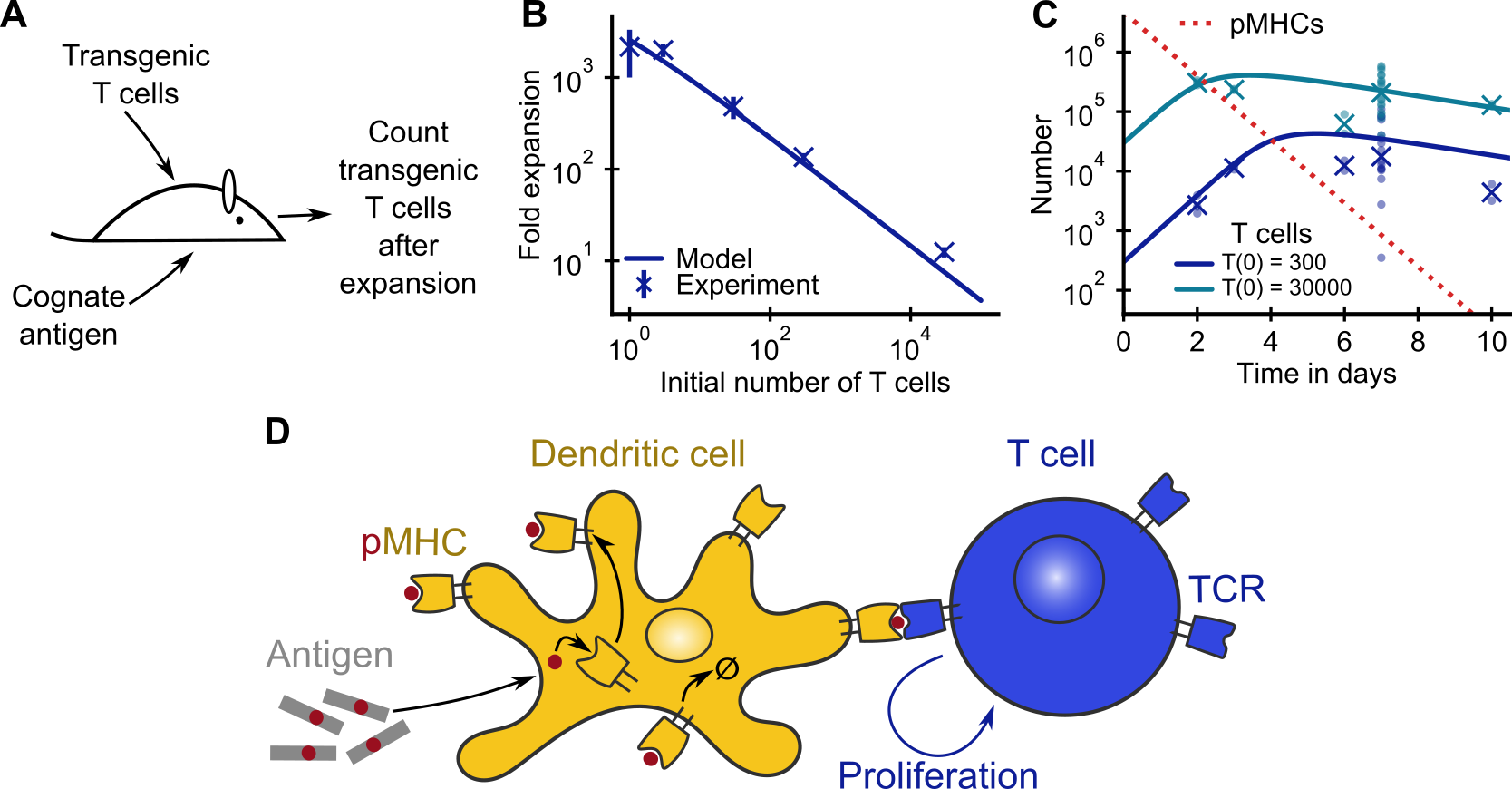
This repository contains the source code associated with the manuscript
Mayer, Zhang, Perelson, Wingreen: [Regulation of T cell expansion by antigen presentation dynamics](https://doi.org/10.1073/pnas.1812800116), PNAS 2019
It allows reproduction of all numerical results reported in the manuscript.
## Installation requirements
The code uses Python 3.6+.
A number of standard scientific python packages are needed for the numerical simulations and visualizations. An easy way to install all of these is to install a Python distribution such as [Anaconda](https://www.continuum.io/downloads).
- [numpy](http://github.com/numpy/numpy/)
- [scipy](https://github.com/scipy/scipy)
- [pandas](http://github.com/pydata/pandas)
- [matplotlib](http://github.com/matplotlib/matplotlib)
Additionally the code also relies on this package:
- [projgrad](https://github.com/andim/projgrad)
## Structure/running the code
Every folder contains a file `plot.py` which needs to be run to produce the figures. For a number of figures cosmetic changes were done in inkscape as a postprocessing step. In these cases the figures will not be reproduced precisely. To help reuse the final edited figures are provided in png/svg format.
## Contact
If you run into any difficulties running the code, feel free to contact us at `andimscience@gmail.com`.
## License
The source code is freely available under an MIT license. The plots are licensed under a Creative commons attributions license (CC-BY).