https://github.com/anselmoo/tanabesugano
A python-solver for Tanabe-Sugano and energy-correlation diagrams
https://github.com/anselmoo/tanabesugano
absorption-spectra complex-ions d5 energy-correlation-diagrams python python3 science simulation spectroscopy tanabe-sugano uvvis
Last synced: 7 months ago
JSON representation
A python-solver for Tanabe-Sugano and energy-correlation diagrams
- Host: GitHub
- URL: https://github.com/anselmoo/tanabesugano
- Owner: Anselmoo
- License: mit
- Created: 2019-09-06T17:58:21.000Z (about 6 years ago)
- Default Branch: master
- Last Pushed: 2025-02-03T21:25:48.000Z (8 months ago)
- Last Synced: 2025-03-15T01:04:53.695Z (7 months ago)
- Topics: absorption-spectra, complex-ions, d5, energy-correlation-diagrams, python, python3, science, simulation, spectroscopy, tanabe-sugano, uvvis
- Language: HTML
- Homepage:
- Size: 23.5 MB
- Stars: 15
- Watchers: 3
- Forks: 2
- Open Issues: 9
-
Metadata Files:
- Readme: README.md
- Changelog: CHANGELOG.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
- Citation: CITATION.cff
Awesome Lists containing this project
README
[](https://github.com/Anselmoo/TanabeSugano/actions/workflows/python-package.yml)
[](https://python-poetry.org/)
[](https://www.codefactor.io/repository/github/anselmoo/tanabesugano)
[](https://zenodo.org/badge/latestdoi/206847682)
[](https://github.com/Anselmoo/TanabeSugano/blob/master/LICENSE)
[](https://github.com/Anselmoo/TanabeSugano/releases)
[](https://pypi.org/project/TanabeSugano/)
[](https://pypi.org/project/TanabeSugano/)
[](https://pepy.tech/project/tanabesugano)
[](https://colab.research.google.com/github/Anselmoo/TanabeSugano/blob/master/Tanabe_Sugano.ipynb)
# TanabeSugano
A python-based Eigensolver for Tanabe-Sugano- & Energy-Correlation-Diagrams based on the original three proposed studies of *Yukito Tanabe and Satoru Sugano* for d2-d8 transition metal ions:
1. On the Absorption Spectra of Complex Ions. I
**Yukito Tanabe, Satoru Sugano**
*Journal of the Physical Society of Japan*, 9, 753-766 (1954)
**DOI:** 10.1143/JPSJ.9.753
https://journals.jps.jp/doi/10.1143/JPSJ.9.753
2. On the Absorption Spectra of Complex Ions II
**Yukito Tanabe, Satoru Sugano**
*Journal of the Physical Society of Japan*, 9, 766-779 (1954)
**DOI:** 10.1143/JPSJ.9.766
https://journals.jps.jp/doi/10.1143/JPSJ.9.766
3. On the Absorption Spectra of Complex Ions, III The Calculation of the Crystalline Field Strength
**Yukito Tanabe, Satoru Sugano**
*Journal of the Physical Society of Japan*, 11, 864-877 (1956)
**DOI:** 10.1143/JPSJ.11.864
https://journals.jps.jp/doi/10.1143/JPSJ.11.864
It provides:
- Tanabe-Sugano- & Energy-Correlation-Diagrams plotted via `matplotlib`
- Tanabe-Sugano- & Energy-Correlation-Diagrams exported as `txt`-file
- Atomic-Termsymbols and their **Eigen-Energies** for a given 10Dq and oxidation state as exported table via `prettytable`
- Set-up individually **C/B**-ratios
- Working with Slater-Condon-Parameters **F2, F4** instead of Racah-Parameters **B, C**
- Export of the **Tanabe-Sugano-Diagram** as a `html`-file via `plotly` for interactive use
The **TanabeSugano**-application can be installed and run:
```console
# via PyPi
pip install TanabeSugano
# via pip+git
pip git+https://github.com/Anselmoo/TanabeSugano.git
# locally
python setup.py install
tanabesugano
# for plotly-export
pip install TanabeSugano[plotly]
```
The options for the **TanabeSugano**-application are:
```console
tanabesugano --help
usage: __main__.py [-h] [-d D] [-Dq DQ] [-cut CUT] [-B B B] [-C C C] [-n N]
[-ndisp] [-ntxt] [-slater]
optional arguments:
-h, --help show this help message and exit
-d D Number of unpaired electrons (default d5)
-Dq DQ 10Dq crystal field splitting (default 10Dq = 8065 cm-)
-cut CUT 10Dq crystal field splitting (default 10Dq = 8065 cm-)
-B B B Racah Parameter B and the corresponding reduction (default B = 860 cm- * 1.)
-C C C Racah Parameter C and the corresponding reduction (default C = 4.477*860 cm- * 1.)
-n N Number of roots (default nroots = 500)
-ndisp Plot TS-diagram (default = on)
-ntxt Save TS-diagram and dd energies (default = on)
-slater Using Slater-Condon F2,F4 parameter instead Racah-Parameter B,C (default = off)
-v, --version Print version number and exit
-html Save TS-diagram and dd energies (default = on)
```
**Reference-Example** for d6 for *B = 860 cm-* and *C = 3850 cm-* as regular `matplotlib`-plot:

**Reference-Example** for d6 for *F2 = 1065 cm-* and *F4 = 5120 cm-* as interactive `plotly`-plot:
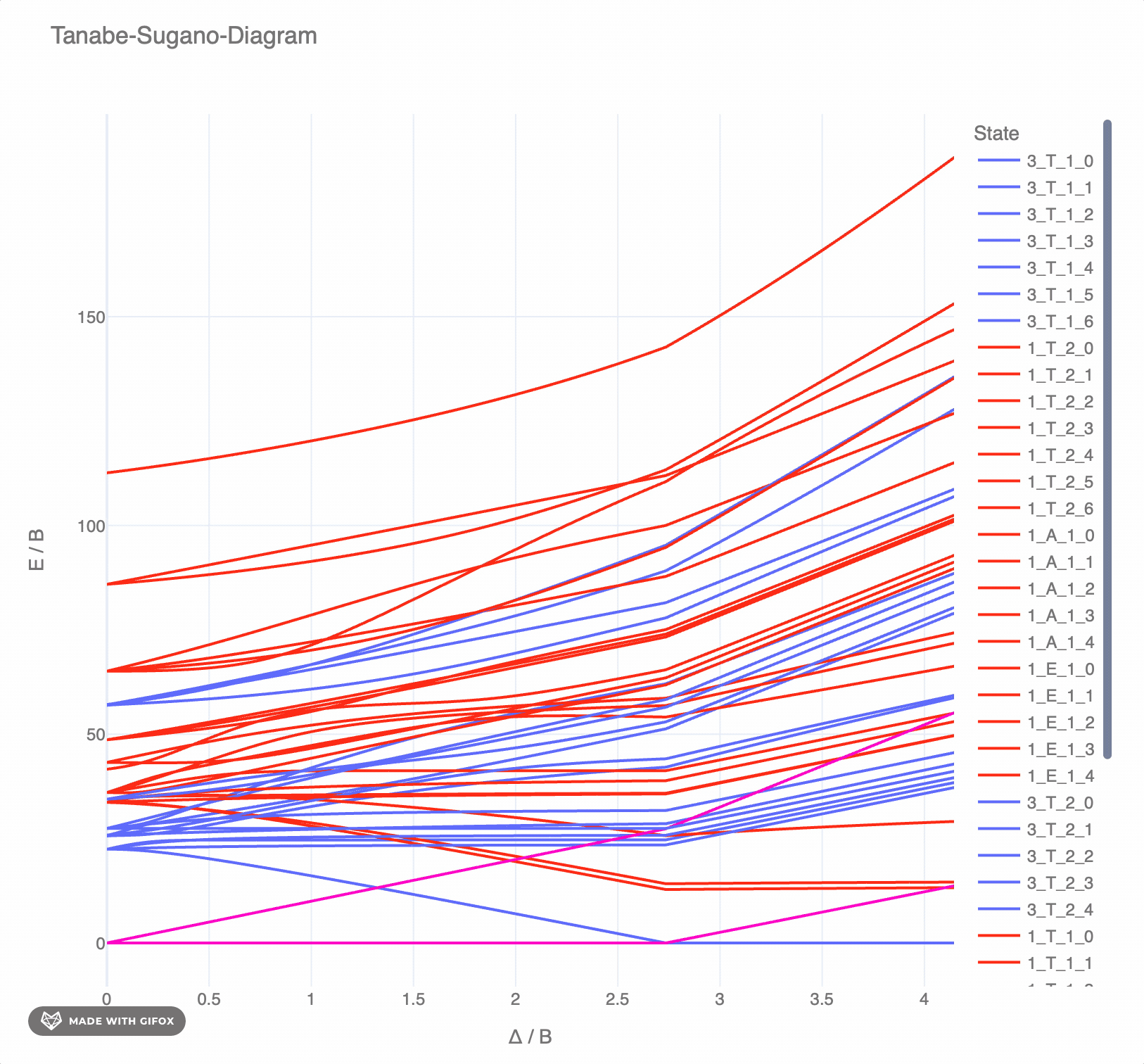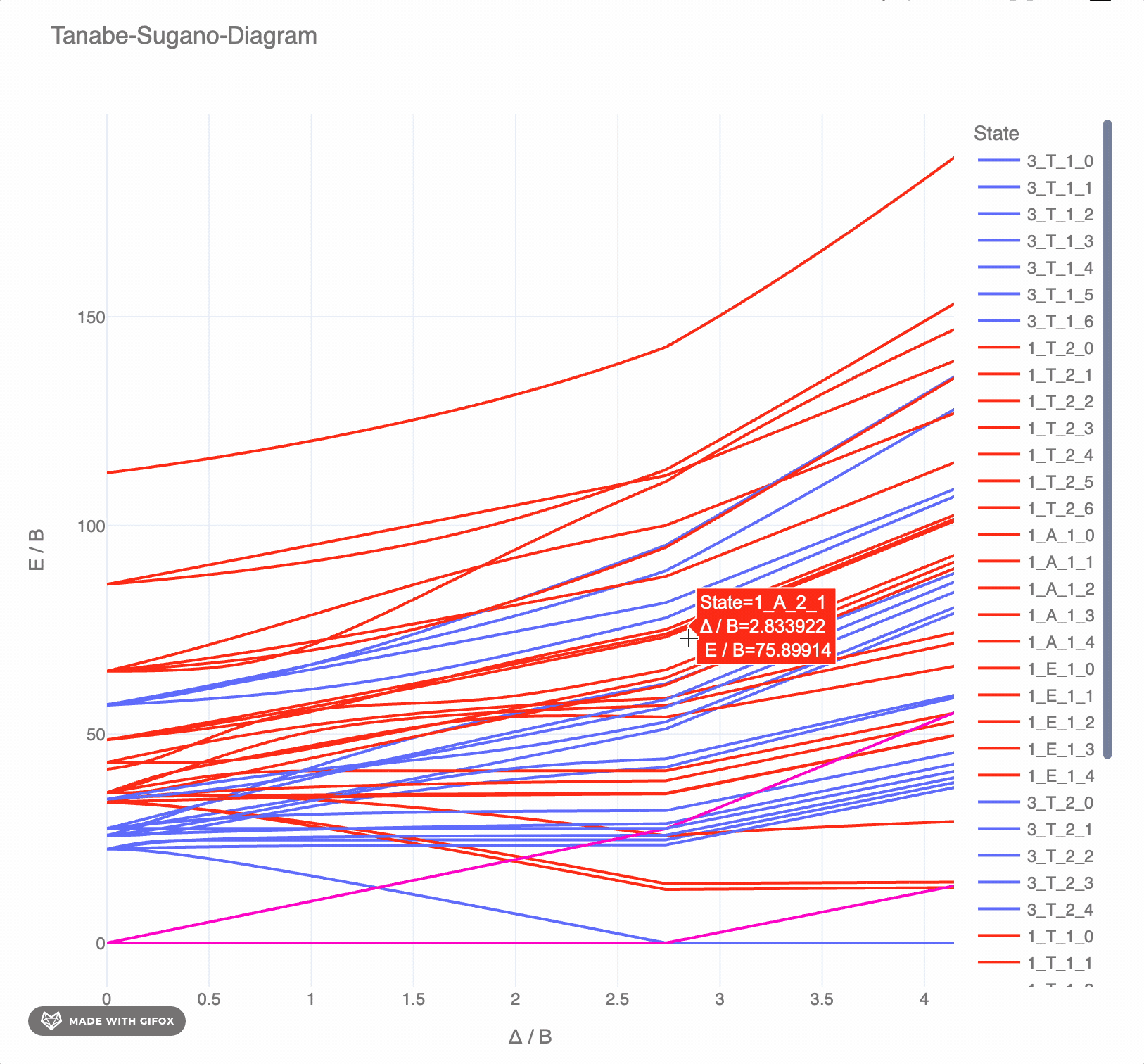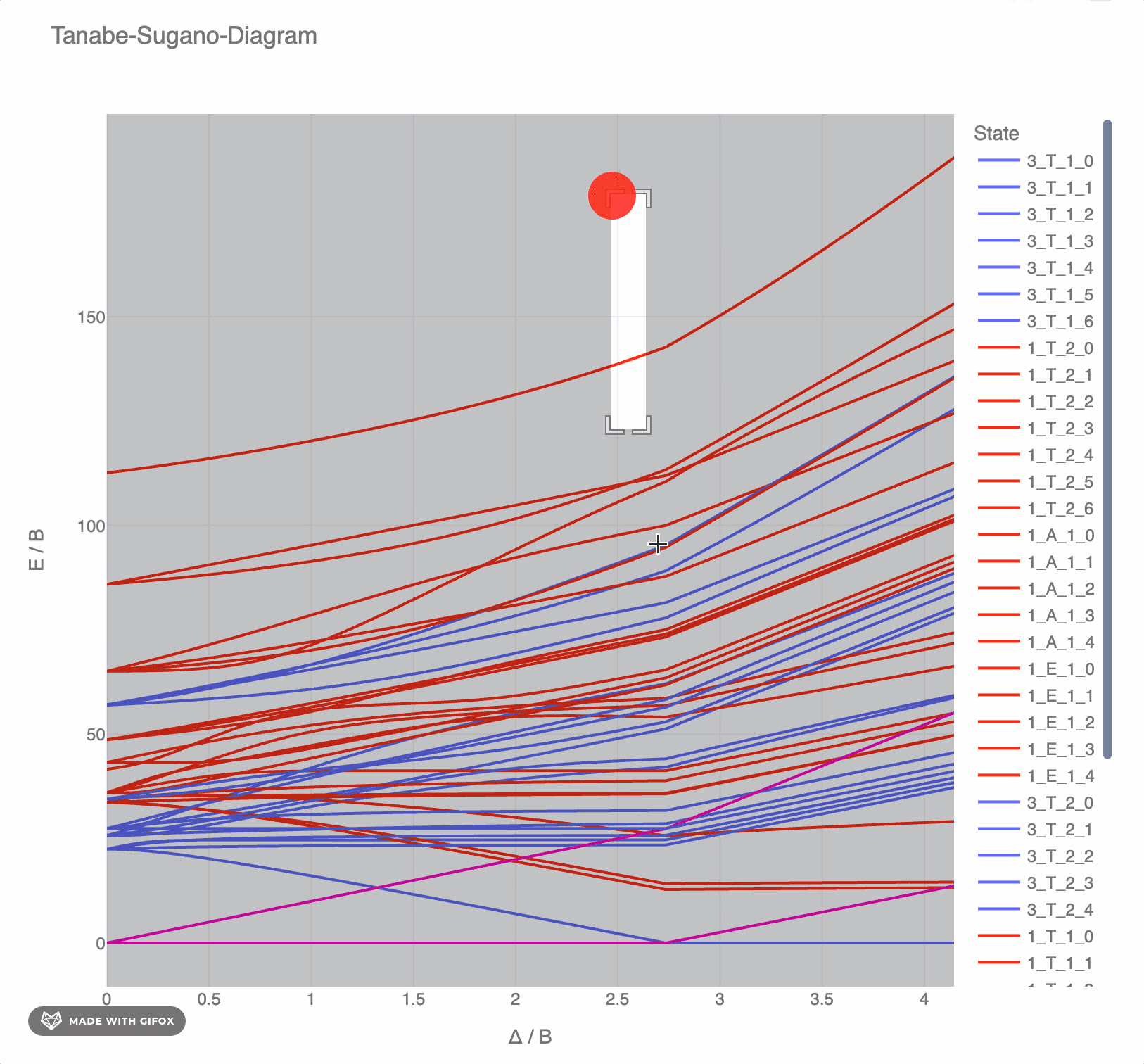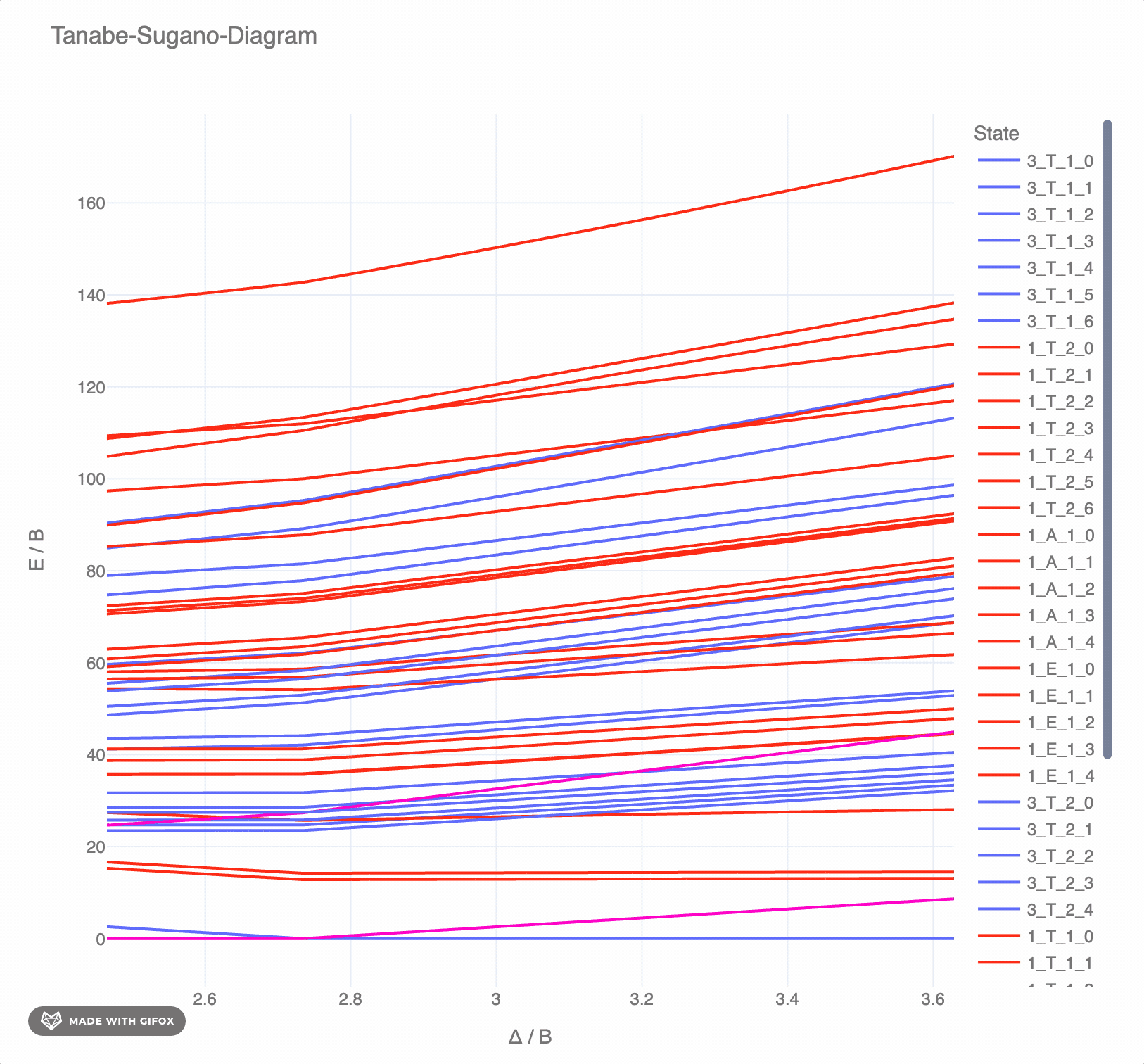
## Test the TS-diagrams interactively in Google Colab
[](https://colab.research.google.com/github/Anselmoo/TanabeSugano/blob/master/Tanabe_Sugano.ipynb)