https://github.com/antonior92/automatic-ecg-diagnosis
Scripts and modules for training and testing neural network for ECG automatic classification. Companion code to the paper "Automatic diagnosis of the 12-lead ECG using a deep neural network".
https://github.com/antonior92/automatic-ecg-diagnosis
atrial-fibrillation atrial-fibrillation-detection convolutional-neural-networks deep-learning ecg ecg-classification ecg-signal
Last synced: 3 months ago
JSON representation
Scripts and modules for training and testing neural network for ECG automatic classification. Companion code to the paper "Automatic diagnosis of the 12-lead ECG using a deep neural network".
- Host: GitHub
- URL: https://github.com/antonior92/automatic-ecg-diagnosis
- Owner: antonior92
- License: mit
- Created: 2020-01-17T19:53:47.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2023-03-25T00:40:18.000Z (about 2 years ago)
- Last Synced: 2025-03-22T18:03:05.750Z (3 months ago)
- Topics: atrial-fibrillation, atrial-fibrillation-detection, convolutional-neural-networks, deep-learning, ecg, ecg-classification, ecg-signal
- Language: Python
- Homepage: https://www.nature.com/articles/s41467-020-15432-4
- Size: 703 KB
- Stars: 378
- Watchers: 8
- Forks: 125
- Open Issues: 5
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Automatic ECG diagnosis using a deep neural network
Scripts and modules for training and testing deep neural networks for ECG automatic classification.
Companion code to the paper "Automatic diagnosis of the 12-lead ECG using a deep neural network".
https://www.nature.com/articles/s41467-020-15432-4.
--------
Citation:
```
Ribeiro, A.H., Ribeiro, M.H., Paixão, G.M.M. et al. Automatic diagnosis of the 12-lead ECG using a deep neural network.
Nat Commun 11, 1760 (2020). https://doi.org/10.1038/s41467-020-15432-4
```
Bibtex:
```
@article{ribeiro_automatic_2020,
title = {Automatic Diagnosis of the 12-Lead {{ECG}} Using a Deep Neural Network},
author = {Ribeiro, Ant{\^o}nio H. and Ribeiro, Manoel Horta and Paix{\~a}o, Gabriela M. M. and Oliveira, Derick M. and Gomes, Paulo R. and Canazart, J{\'e}ssica A. and Ferreira, Milton P. S. and Andersson, Carl R. and Macfarlane, Peter W. and Meira Jr., Wagner and Sch{\"o}n, Thomas B. and Ribeiro, Antonio Luiz P.},
year = {2020},
volume = {11},
pages = {1760},
doi = {https://doi.org/10.1038/s41467-020-15432-4},
journal = {Nature Communications},
number = {1}
}
```
-----
## Requirements
This code was tested on Python 3 with Tensorflow `2.2`. There is an older branch ([`tensorflow-v1`](https://github.com/antonior92/automatic-ecg-diagnosis/tree/tensorflow-v1)) that
contain the code implementation for Tensorflow `1.15`.
**For pytorch users:** If you are interested in a pytorch implementation, take a look in the repository: https://github.com/antonior92/ecg-age-prediction.
There we provide a implementation in PyTorch of the same resnet-based model. The problem there is the age prediction from the ECG, nontheless simple modifications should suffice for dealing with abnormality classification.
## Model
The model used in the paper is a residual neural. The neural network architecture implementation in Keras is available in ``model.py``. To print a summary of the model layers run:
```bash
$ python model.py
```
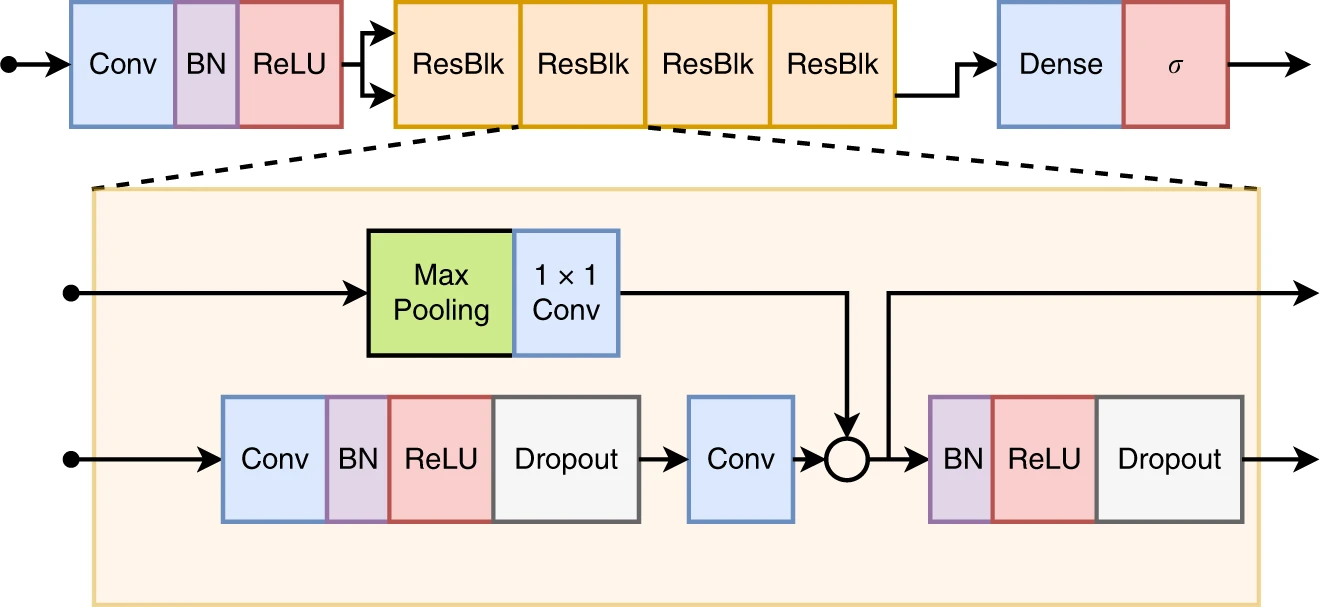
The model receives an input tensor with dimension `(N, 4096, 12)`, and returns an output tensor with dimension `(N, 6)`,
for which `N` is the batch size.
The model can be trained using the script `train.py`. Alternatively,
pre-trained weighs for the models described in the paper are also
available in: https://doi.org/10.5281/zenodo.3625017. Or in the mirror dropbox
link [here](https://www.dropbox.com/s/5ar6j8u9v9a0rmh/model.zip?dl=0).
Using the command line, the weights can be downloaded using
```
wget https://www.dropbox.com/s/5ar6j8u9v9a0rmh/model.zip?dl=0 -O model.zip
unzip model.zip
```
- **input**: `shape = (N, 4096, 12)`. The input tensor should contain the `4096` points of the ECG tracings
sampled at `400Hz` (i.e., a signal of approximately 10 seconds). Both in the training and in the test set, when the
signal was not long enough, we filled the signal with zeros, so 4096 points were attained. The last dimension of the
tensor contains points of the 12 different leads. The leads are ordered in the following order:
`{DI, DII, DIII, AVR, AVL, AVF, V1, V2, V3, V4, V5, V6}`. All signal are represented as
32 bits floating point numbers at the scale 1e-4V: so if the signal is in V it should be multiplied by
1000 before feeding it to the neural network model.
- **output**: `shape = (N, 6)`. Each entry contains a probability between 0 and 1, and can be understood as the
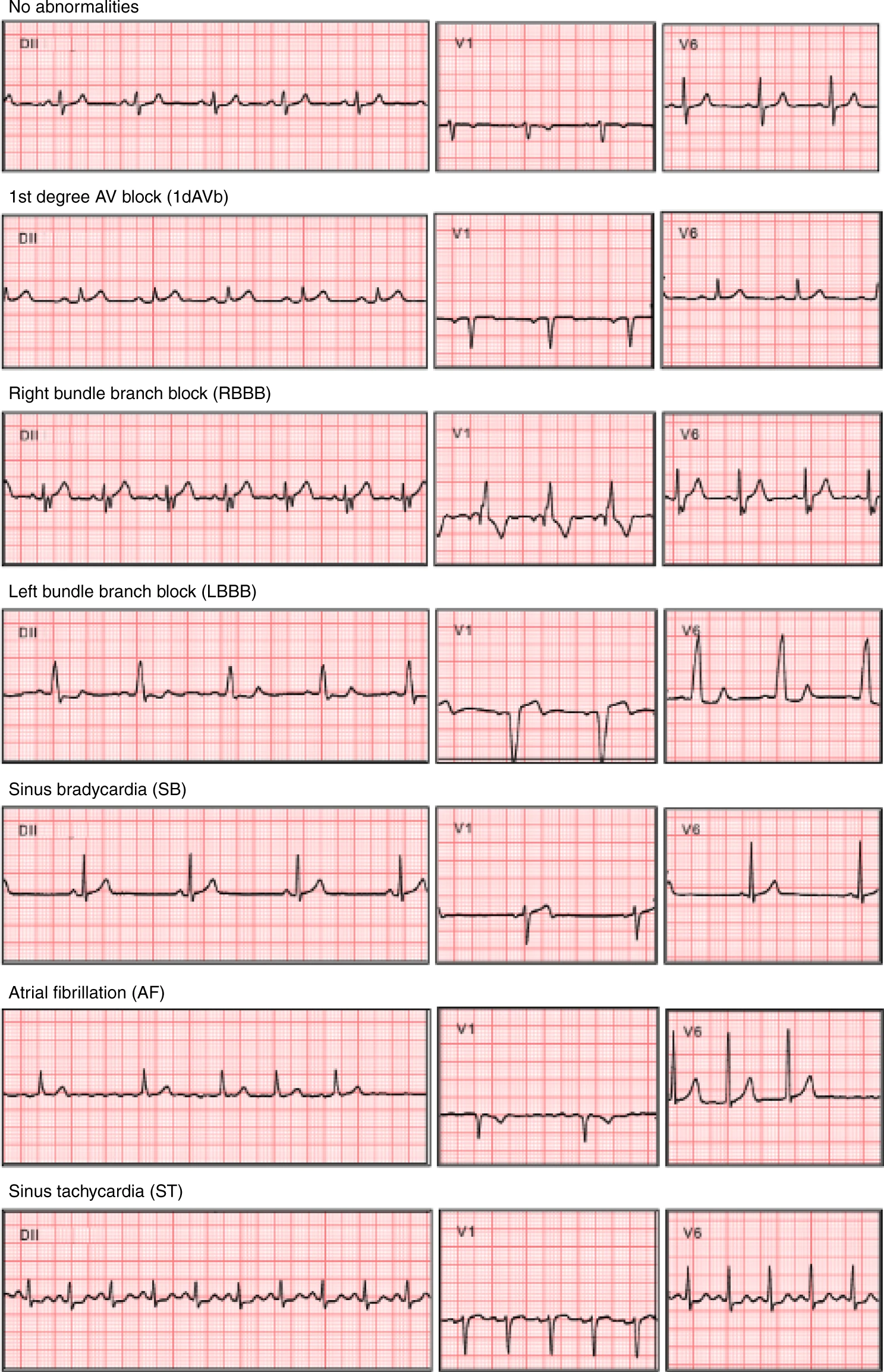
probability of a given abnormality to be present. The abnormalities it predicts are **(in that order)**: 1st degree AV block(1dAVb),
right bundle branch block (RBBB), left bundle branch block (LBBB), sinus bradycardia (SB), atrial fibrillation (AF),
sinus tachycardia (ST). The abnormalities are not mutually exclusive, so the probabilities do not necessarily
sum to one.
## Datasets
- The testing dataset described in the paper can be downloaded in:
[doi: 10.5281/zenodo.3625006](https://doi.org/10.5281/zenodo.3625006). There is also a mirror
dropbox link [here](https://www.dropbox.com/s/p3vd3plcbu9sf1o/data.zip?dl=0).
Using the command line:
```
wget https://www.dropbox.com/s/p3vd3plcbu9sf1o/data.zip?dl=0 -O data.zip
unzip data.zip
```
- Part of the training data (the CODE-15\% dataset) is openly available in: [doi: 10.5281/zenodo.4916206 ](https://doi.org/10.5281/zenodo.4916206).
- The full CODE dataset that was used for training is available upon request for research purposes: [doi: 10.17044/scilifelab.15169716](https://doi.org/10.17044/scilifelab.15169716)
## Scripts
- ``train.py``: Script for training the neural network. To train the neural network run:
```bash
$ python train.py PATH_TO_HDF5 PATH_TO_CSV
```
Pre-trained models obtained using such script can be downloaded from [here](https://doi.org/10.5281/zenodo.3625017)
- ``predict.py``: Script for generating the neural network predictions on a given dataset.
```bash
$ python predict.py PATH_TO_HDF5_ECG_TRACINGS PATH_TO_MODEL --ouput_file PATH_TO_OUTPUT_FILE
```
The folder `./dnn_predicts` contain the output obtained by applying this script to the models available in
[here](https://doi.org/10.5281/zenodo.3625017) to make the predictions on tracings from
[this test dataset](https://doi.org/10.5281/zenodo.3625006).
- ``generate_figures_and_tables.py``: Generate figures and tables from the paper "Automatic Diagnosis o
the Short-Duration12-Lead ECG using a Deep Neural Network". Make sure to execute the script from the root folder,
so all relative paths are correct. So first run:
```
$ cd /path/to/automatic-ecg-diagnosis
```
Then the script
```bash
$ python generate_figures_and_tables.py
```
It should generate the tables and figure in the folder `outputs/`
- ``model.py``: Auxiliary module that defines the architecture of the deep neural network.
To print a summary of the model layers run:
```bash
$ python model.py
```