Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/aramis-lab/clinica
Software platform for clinical neuroimaging studies
https://github.com/aramis-lab/clinica
ants bids-format brainweb freesurfer fsl machine-learning mrtrix3 neuroimaging neuroscience python scikit-learn spm
Last synced: 2 months ago
JSON representation
Software platform for clinical neuroimaging studies
- Host: GitHub
- URL: https://github.com/aramis-lab/clinica
- Owner: aramis-lab
- License: other
- Created: 2019-05-17T13:50:31.000Z (over 5 years ago)
- Default Branch: dev
- Last Pushed: 2024-10-25T07:01:31.000Z (3 months ago)
- Last Synced: 2024-10-25T10:23:01.879Z (3 months ago)
- Topics: ants, bids-format, brainweb, freesurfer, fsl, machine-learning, mrtrix3, neuroimaging, neuroscience, python, scikit-learn, spm
- Language: Python
- Homepage: http://www.clinica.run/
- Size: 34.5 MB
- Stars: 222
- Watchers: 13
- Forks: 75
- Open Issues: 78
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE.txt
- Citation: CITATION.cff
Awesome Lists containing this project
README

Clinica
Software platform for clinical neuroimaging studies
Homepage |
Documentation |
Paper |
Forum |
See also:
AD-ML,
AD-DL,
ClinicaDL
## About The Project
Clinica is a software platform for clinical research studies involving patients
with neurological and psychiatric diseases and the acquisition of multimodal
data (neuroimaging, clinical and cognitive evaluations, genetics...),
most often with longitudinal follow-up.
Clinica is command-line driven and written in Python.
It uses the [Nipype](https://nipype.readthedocs.io/) system for pipelining and combines
widely-used software packages for neuroimaging data analysis
([ANTs](http://stnava.github.io/ANTs/),
[FreeSurfer](https://surfer.nmr.mgh.harvard.edu/),
[FSL](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki),
[MRtrix](https://www.mrtrix.org/),
[PETPVC](https://github.com/UCL/PETPVC),
[SPM](https://www.fil.ion.ucl.ac.uk/spm/)), machine learning
([Scikit-learn](https://scikit-learn.org/stable/)) and the [BIDS
standard](http://bids-specification.readthedocs.io/) for data organization.
Clinica provides tools to convert publicly available neuroimaging datasets into
BIDS, namely:
- [ADNI: Alzheimer’s Disease Neuroimaging Initiative](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Converters/ADNI2BIDS/)
- [AIBL: Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Converters/AIBL2BIDS/)
- [HABS: Harvard Aging Brain Study](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Converters/HABS2BIDS/)
- [NIFD: Neuroimaging in Frontotemporal Dementia](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Converters/NIFD2BIDS/)
- [OASIS: Open Access Series of Imaging Studies](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Converters/OASIS2BIDS/)
- [OASIS-3: Longitudinal Neuroimaging, Clinical, and Cognitive Dataset for Normal Aging and Alzheimer’s Disease](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Converters/OASIS3TOBIDS/)
Clinica can process any BIDS-compliant dataset with a set of complex processing
pipelines involving different software packages for the analysis of
neuroimaging data (T1-weighted MRI, diffusion MRI and PET data).
It also provides integration between feature extraction and statistics, machine
learning or deep learning.
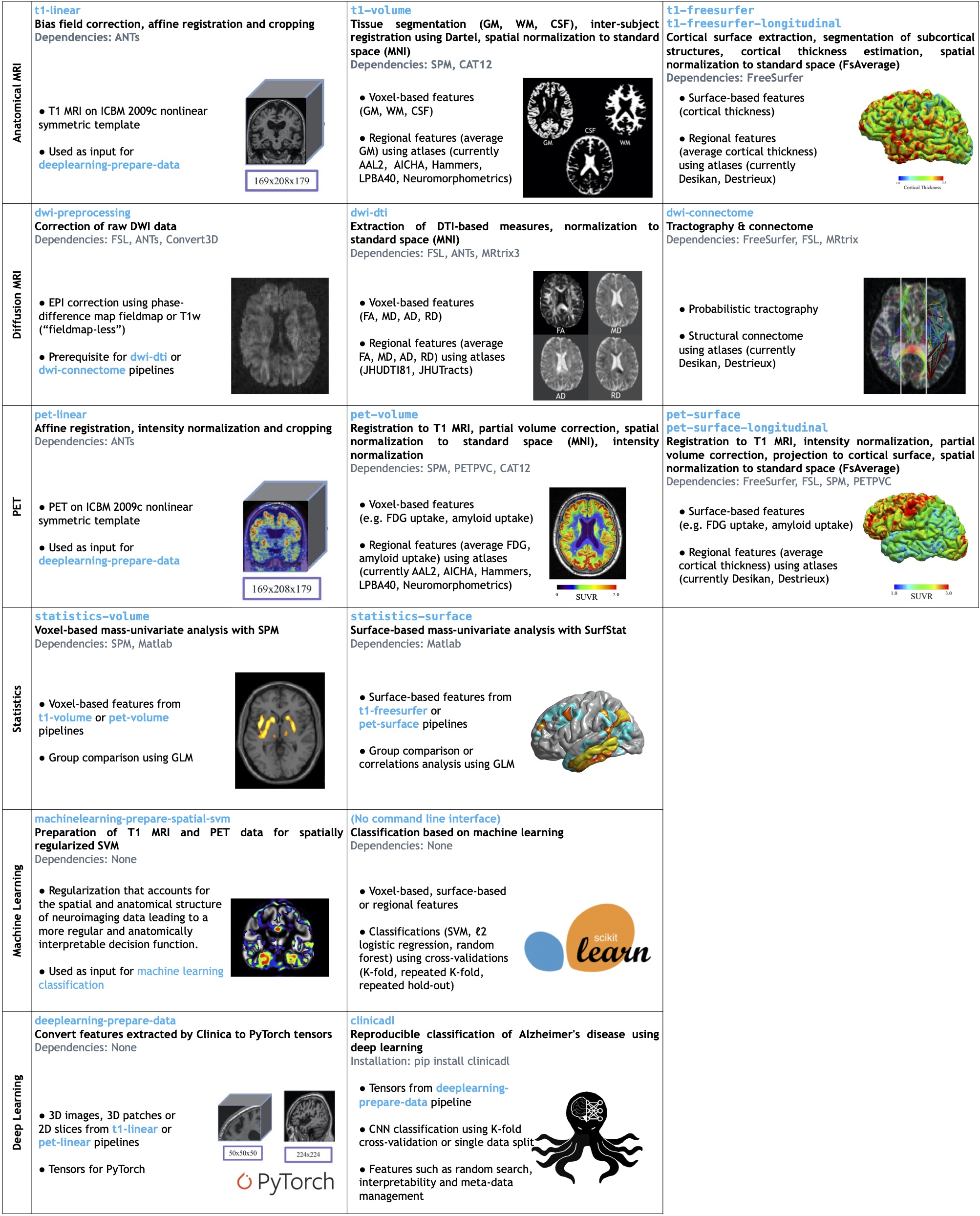
Clinica is also showcased as a framework for the reproducible classification of
Alzheimer's disease using
[machine learning](https://github.com/aramis-lab/AD-ML) and
[deep learning](https://github.com/aramis-lab/clinicadl).
## Getting Started
> Full instructions for installation and additional information can be found in
the [user documentation](https://aramislab.paris.inria.fr/clinica/docs/public/latest/).
### Using pipx (recommended)
Clinica can be easily installed and updated using [pipx](https://pypa.github.io/pipx/).
```console
pipx install clinica
```
### Using pip
```console
pip install clinica
```
### Using Conda
Clinica relies on multiple third-party tools to perform processing.
An environment file is provided in this repository
to facilitate their installation in a [Conda](https://docs.conda.io/en/latest/miniconda.html) environment:
```console
git clone https://github.com/aramis-lab/clinica && cd clinica
conda env create
conda activate clinica
```
After activation, use `pip` to install Clinica.
### Additional dependencies (required)
Depending on the pipeline that you want to use, you need to install pipeline-specific interfaces.
Some of which uses a different runtime or use incompatible licensing terms, which prevent their distribution alongside Clinica.
Not all the dependencies are necessary to run Clinica.
Please refer to this [page](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Third-party/)
to determine which third-party libraries you need to install.
## Example
Diagram illustrating the Clinica pipelines involved when performing a group
comparison of FDG PET data projected on the cortical surface between patients
with Alzheimer's disease and healthy controls from the ADNI database:
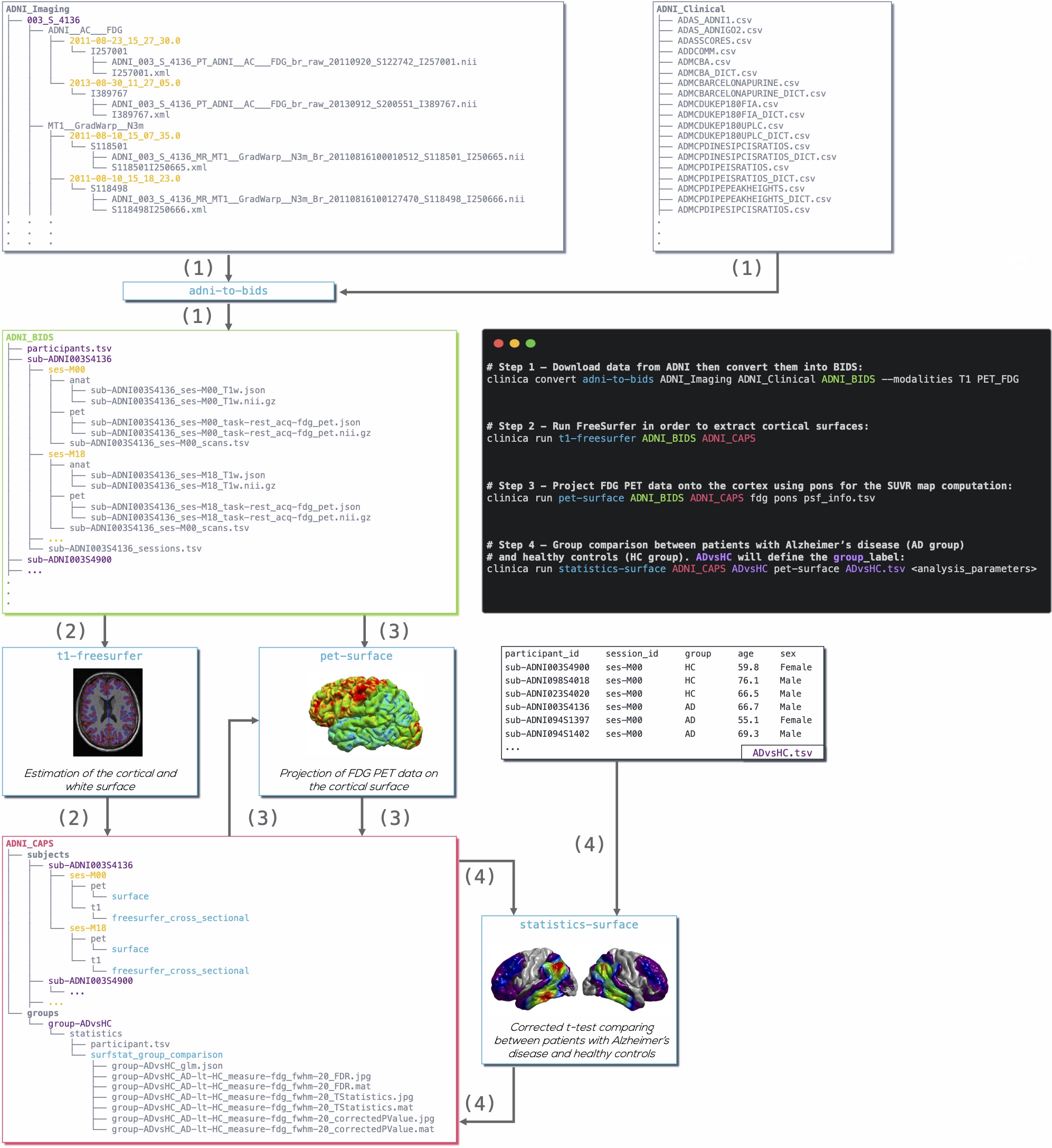
1. Clinical and neuroimaging data are downloaded from the ADNI website and data
are converted into BIDS with the [`adni-to-bids`
converter](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Converters/ADNI2BIDS/).
2. Estimation of the cortical and white surface is then produced by the
[`t1-freesurfer`
pipeline](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Pipelines/T1_FreeSurfer/).
3. FDG PET data can be projected on the subject’s cortical surface and
normalized to the FsAverage template from FreeSurfer using the
[`pet-surface` pipeline](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Pipelines/PET_Surface/).
4. TSV file with demographic information of the population studied is given to
the [`statistics-surface`
pipeline](https://aramislab.paris.inria.fr/clinica/docs/public/latest/Pipelines/Stats_Surface/) to generate
the results of the group comparison.
> For more examples and details, please refer to the
> [Documentation](https://aramislab.paris.inria.fr/clinica/docs/public/latest/).
## Support
- Check for [past answers](https://groups.google.com/forum/#!forum/clinica-user) in the old Clinica Google Group
- Start a [discussion](https://github.com/aramis-lab/clinica/discussions) on Github
- Report an [issue](https://github.com/aramis-lab/clinica/issues) on GitHub
## Contributing
We encourage you to contribute to Clinica!
Please check out the [Contributing to Clinica guide](CONTRIBUTING.md) for
guidelines about how to proceed. Do not hesitate to ask questions if something
is not clear for you, report an issue, etc.
## License
This software is distributed under the MIT License.
See [license file](https://github.com/aramis-lab/clinica/blob/dev/LICENSE.txt)
for more information.
## Citing us
- Routier, A., Burgos, N., Díaz, M., Bacci, M., Bottani, S., El-Rifai O., Fontanella, S., Gori, P., Guillon, J., Guyot, A., Hassanaly, R., Jacquemont, T., Lu, P., Marcoux, A., Moreau, T., Samper-González, J., Teichmann, M., Thibeau-Sutre, E., Vaillant G., Wen, J., Wild, A., Habert, M.-O., Durrleman, S., and Colliot, O.:
*Clinica: An Open Source Software Platform for Reproducible Clinical Neuroscience Studies* Frontiers in Neuroinformatics, 2021
[doi:10.3389/fninf.2021.689675](https://doi.org/10.3389/fninf.2021.689675)
## Related Repositories
- [AD-DL: Classification of Alzheimer's disease status with convolutional neural networks](https://github.com/aramis-lab/AD-DL).
- [AD-ML: Framework for the reproducible classification of Alzheimer's disease using
machine learning](https://github.com/aramis-lab/AD-ML).
- [ClinicaDL: Framework for the reproducible processing of neuroimaging data with deep learning methods](https://github.com/aramis-lab/clinicadl).


