Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/biapyx/biapy-gui
BiaPy GUI
https://github.com/biapyx/biapy-gui
Last synced: about 21 hours ago
JSON representation
BiaPy GUI
- Host: GitHub
- URL: https://github.com/biapyx/biapy-gui
- Owner: BiaPyX
- License: mit
- Created: 2023-06-22T11:11:49.000Z (over 1 year ago)
- Default Branch: main
- Last Pushed: 2024-05-23T08:16:32.000Z (6 months ago)
- Last Synced: 2024-05-23T08:58:09.139Z (6 months ago)
- Language: Python
- Size: 133 MB
- Stars: 6
- Watchers: 2
- Forks: 0
- Open Issues: 2
-
Metadata Files:
- Readme: README.md
- License: LICENSE.md
Awesome Lists containing this project
README
# Graphical user interface for BiaPy: Bioimage analysis pipelines in Python
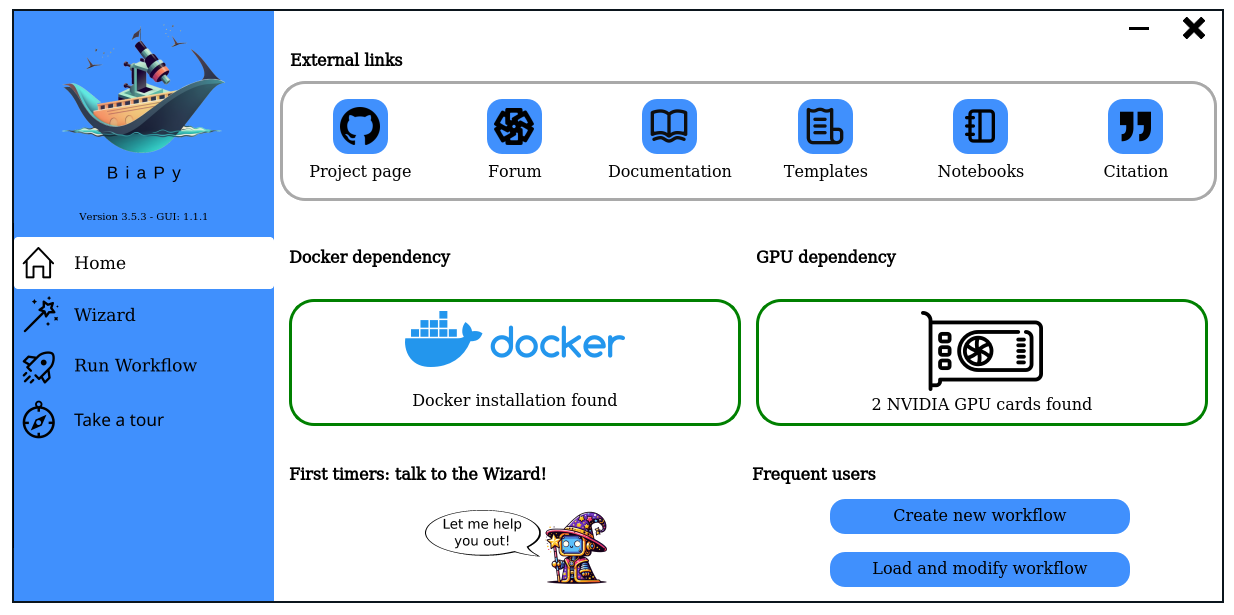
## Download BiaPy GUI for you OS
- [Windows 64-bit](https://drive.google.com/uc?export=download&id=1iV0wzdFhpCpBCBgsameGyT3iFyQ6av5o)
- [Linux 64-bit](https://drive.google.com/uc?export=download&id=13jllkLTR6S3yVZLRdMwhWUu7lq3HyJsD)
- [macOS 64-bit](https://drive.google.com/uc?export=download&id=1fIpj9A8SWIN1fABEUAS--DNhOHzqSL7f)
## BiaPy
[BiaPy](https://biapyx.github.io) is an open source ready-to-use all-in-one library that provides deep-learning workflows for a large variety of bioimage analysis tasks, including 2D and 3D [semantic segmentation](https://biapy.readthedocs.io/en/latest/workflows/semantic_segmentation.html), [instance segmentation](https://biapy.readthedocs.io/en/latest/workflows/instance_segmentation.html), [object detection](https://biapy.readthedocs.io/en/latest/workflows/detection.html), [image denoising](https://biapy.readthedocs.io/en/latest/workflows/denoising.html), [single image super-resolution](https://biapy.readthedocs.io/en/latest/workflows/super_resolution.html), [self-supervised learning](https://biapy.readthedocs.io/en/latest/workflows/self_supervision.html) and [image classification](https://biapy.readthedocs.io/en/latest/workflows/classification.html).
BiaPy is a versatile platform designed to accommodate both proficient computer scientists and users less experienced in programming. It offers diverse and user-friendly access points to our workflows.
This repository is actively under development by the Biomedical Computer Vision group at the [University of the Basque Country](https://www.ehu.eus/en/en-home) and the [Donostia International Physics Center](http://dipc.ehu.es/).

## Description video
Find a comprehensive overview of BiaPy and its functionality in the following videos:
| [](https://www.youtube.com/watch?v=Gnm-VsZQ6Cc "BiaPy: a ready-to-use library for Bioimage Analysis Pipelines")
BiaPy history and GUI demo at [RTmfm](https://rtmfm.cnrs.fr/en/) by Ignacio Arganda-Carreras and Daniel Franco-Barranco. |
|-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
| [](https://www.youtube.com/watch?v=6eYtX-ySpc0 "BiaPy: a ready-to-use library for Bioimage Analysis Pipelines")
BiaPy presentation at [Virtual Pub of Euro-BioImaging](https://www.eurobioimaging.eu/) by Ignacio Arganda-Carreras. | | | |
## For developers (through console):
Download the repository first:
```shell
git clone https://github.com/BiaPyX/BiaPy-GUI.git
cd BiaPy-GUI
```
This will create a folder called ``BiaPy-GUI`` that contains all the files of the repository. Then you need to create a conda environment and install the dependencies:
```shell
conda create -n biapy-gui python=3.10.10
conda activate biapy-gui
pip install -r requirements.txt
```
After that simply run the ``main.py`` :
```shell
python main.py
```
To create the binary files:
```shell
pyinstaller -F main.py
```
You can also modify the main.spec file (e.g. to add new images) and redo the binary:
```shell
pyinstaller main.spec
```
This will create a ``main.spec`` file and a ``dist`` folder with ``BiaPy`` binary inside it.
## Citation
```
Franco-Barranco, Daniel, et al. "BiaPy: a ready-to-use library for Bioimage Analysis Pipelines."
2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI). IEEE, 2023.
```