https://github.com/bluegreen-labs/geoloctools
A Geolocation Tracking Toolbox
https://github.com/bluegreen-labs/geoloctools
animal-behavior ecology geolocation geolocator logging migration movement-ecology
Last synced: 7 months ago
JSON representation
A Geolocation Tracking Toolbox
- Host: GitHub
- URL: https://github.com/bluegreen-labs/geoloctools
- Owner: bluegreen-labs
- License: other
- Created: 2022-01-17T22:40:55.000Z (over 3 years ago)
- Default Branch: main
- Last Pushed: 2022-12-31T13:25:08.000Z (almost 3 years ago)
- Last Synced: 2025-01-24T08:11:25.855Z (9 months ago)
- Topics: animal-behavior, ecology, geolocation, geolocator, logging, migration, movement-ecology
- Language: R
- Homepage: https://bluegreen-labs.github.io/geoloctools/
- Size: 181 MB
- Stars: 0
- Watchers: 1
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# GeoLocTools 
A set of functions to pre-process geolocation tracking data. This includes
both light loggers, GPS loggers of all kinds, and tries to ensure
interchangeability between various devices and formats
for synthesis and analysis. Currently the set of functions is limited but we
are sure this set will grow over time, due to personal use or user
contributions.
Due to the inclusion of a high resolution geoid map the package is fairly large
(97 MB). This is less than ideal, yet functional. A solution will be sought for
this situation to be more CRAN compliant.
Warning: this package is made on personal request and therefore is a temporary
convenience tool. Although the intention is to solidify some of this more
permanently, when and how is to be seen. The package might therefore change
name and function at any time. Do not rely on this code operationally in
an online workflow.
## Installation
### development release
To install the development releases of the package run the following
commands:
``` r
if(!require(devtools)){install.packages("devtools")}
devtools::install_github("bluegreen-labs/geoloctools")
library("geoloctools")
```
## Use
### Reading PathTrack position (.pos) files
The `glt_read_pos()` function reads in PathTrack position files into a
data frame. By default the altitudes are reported as values above mean sea
level (corrected from ellipsoide heights to topographic heights by
compensating for [egm2008 one degree geoid model](https://geographiclib.sourceforge.io/1.18/geoid.html)). We do note that due to
the bilinear interpolation instead of cubic interpolation a slight loss in
accuracy is noted. However, with an error of 0.025m this value is well below
the error on commercial animal loggers.
```r
library(geoloctools)
df <- glt_read_pos("your_position_file.pos")
```
### Converting Migrate Technology data to PAMLr formatting
If you've installed the package you can also generate activity and light logging
plot (based upon the code in the `PAMLr` documentation) by converting Migrate
Technology light logger files to the `PAMLr` R package format for further
processing.
```r
library(geoloctools)
library(pamlr)
library(tidyverse)
# load demo data from Migrate Technologies files
PAM <- glt_migtech_pamlr(
system.file(
"extdata",
package = "geoloctools",
mustWork = TRUE),
drift_adj = FALSE
)
# grab one particular logger (subset main list)
PAM_data <- PAM$CC893
# plot activity and light levels as shown in the
# PAMLr documentation
par( mfrow= c(1,2), oma=c(0,2,0,6))
par(mar = c(4,2,4,2))
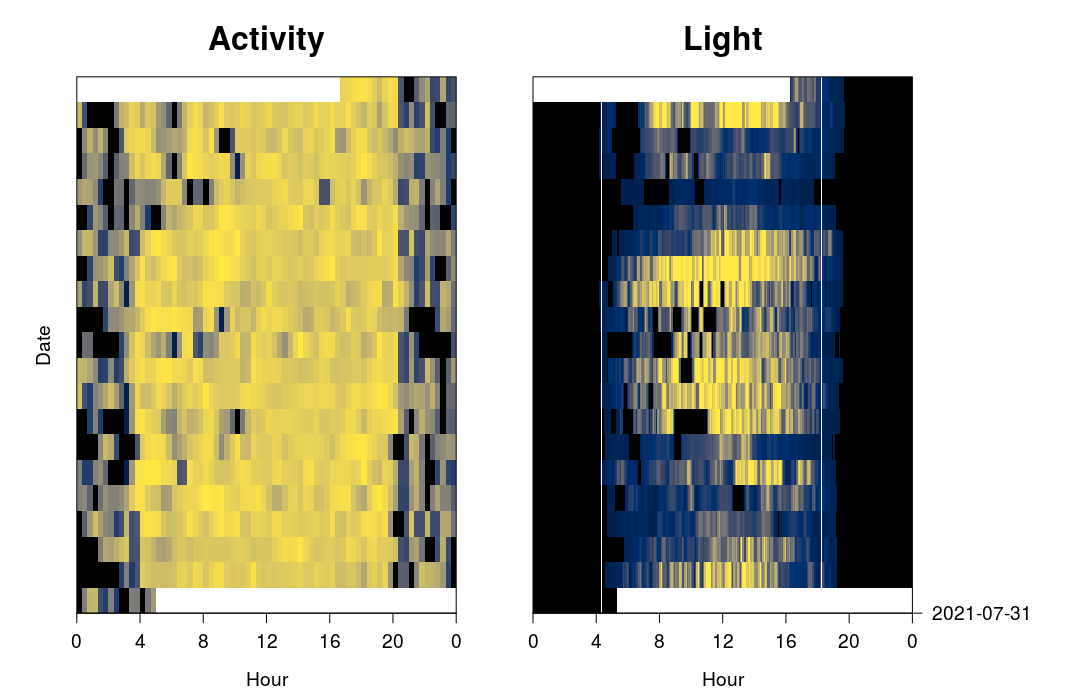
plot_sensorimage(PAM_data$acceleration$date, ploty=FALSE,
log(PAM_data$acceleration$act+0.001), main = "Activity",
col=c("black",viridis::cividis(90)), cex=1.2, cex.main = 2)
plot_sensorimage(PAM_data$light$date, labely=FALSE,
PAM_data$light$obs, main="Light",
col=c("black",viridis::cividis(90)), cex=1.2, cex.main = 2)
# loop over all available items in the list and
# plot them
lapply(PAM, function(PAM_subset){
# plot activity and light levels as shown in the
# PAMLr documentation
par( mfrow= c(1,2), oma=c(0,2,0,6))
par(mar = c(4,2,4,2))
plot_sensorimage(PAM_subset$acceleration$date, ploty=FALSE,
log(PAM_subset$acceleration$act+0.001), main = "Activity",
col=c("black",viridis::cividis(90)), cex=1.2, cex.main = 2)
plot_sensorimage(PAM_subset$light$date, labely=FALSE,
PAM_subset$light$obs, main="Light",
col=c("black",viridis::cividis(90)), cex=1.2, cex.main = 2)
})
```