https://github.com/brainglobe/brainrender
A Python package to visualise neuroanatomical data in atlas space
https://github.com/brainglobe/brainrender
anatomy neuroscience python visualization
Last synced: about 1 month ago
JSON representation
A Python package to visualise neuroanatomical data in atlas space
- Host: GitHub
- URL: https://github.com/brainglobe/brainrender
- Owner: brainglobe
- License: bsd-3-clause
- Created: 2019-09-19T09:27:09.000Z (over 6 years ago)
- Default Branch: main
- Last Pushed: 2025-12-02T16:47:54.000Z (3 months ago)
- Last Synced: 2026-01-04T03:03:12.492Z (about 2 months ago)
- Topics: anatomy, neuroscience, python, visualization
- Language: Python
- Homepage: https://brainglobe.info/documentation/brainrender/index.html
- Size: 271 MB
- Stars: 616
- Watchers: 15
- Forks: 94
- Open Issues: 10
-
Metadata Files:
- Readme: README.md
- License: LICENSE
- Citation: CITATION.cff
Awesome Lists containing this project
- awesome-biological-image-analysis - Brainrender - Python package for the visualization of three dimensional neuro-anatomical data. (Neuroscience)
README
# brainrender
*A user-friendly python library to create high-quality, 3D neuro-anatomical renderings combining data from publicly available brain atlases with user-generated experimental data.*
[](https://pypi.org/project/brainrender)
[](https://pypi.org/project/brainrender)
[](https://github.com/brainglobe/brainrender/actions)
[](https://codecov.io/gh/brainglobe/brainrender)
[](https://pepy.tech/project/brainrender)
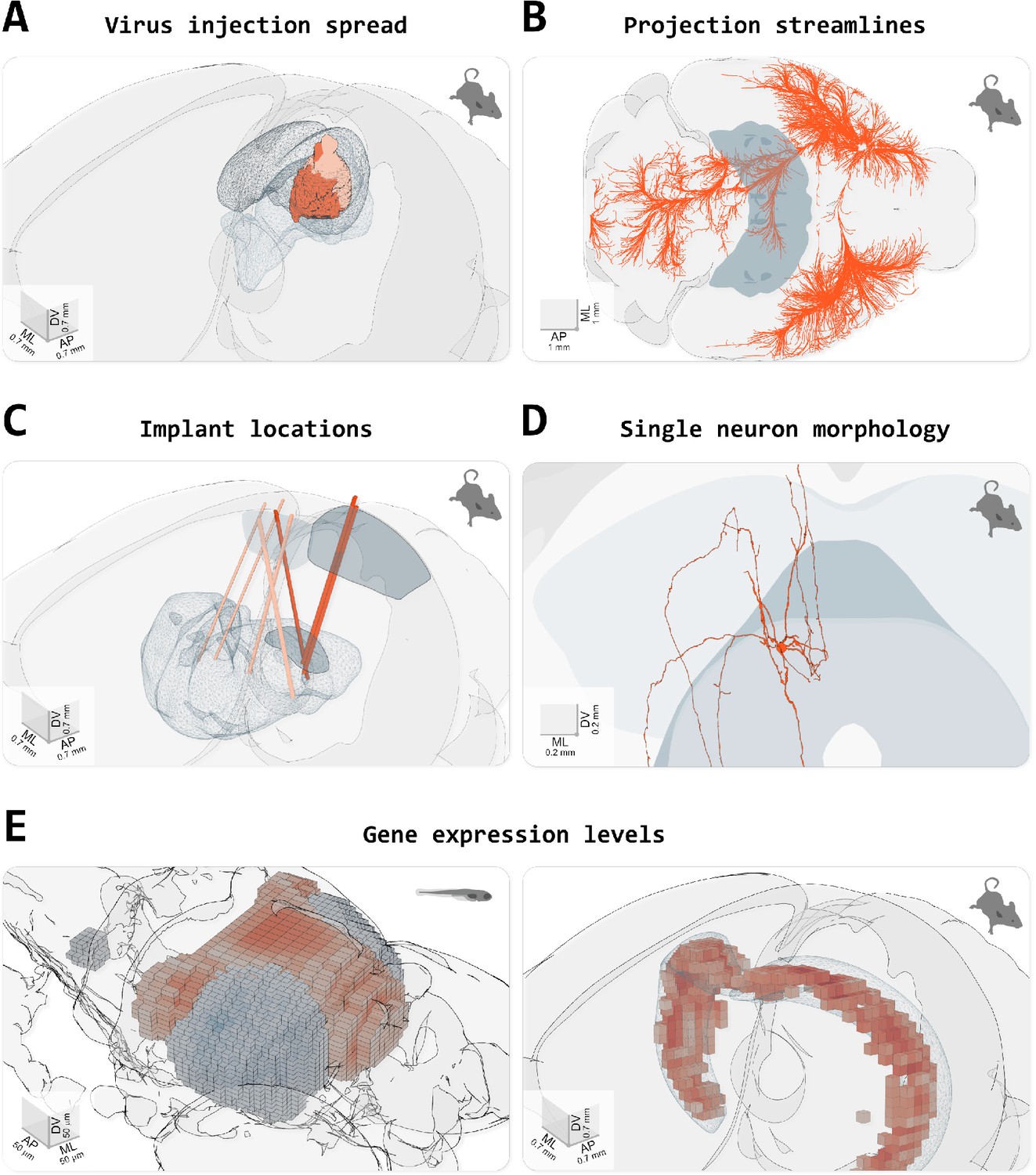
From: Claudi et al. (2021) Visualizing anatomically registered data with brainrender. eLife
## Documentation
brainrender is a project of the BrainGlobe Initiative, which is a collaborative effort to develop a suite of Python-based software tools for computational neuroanatomy. A comprehensive online documentation for brainrender can be found on the BrainGlobe website [here](https://brainglobe.info/documentation/brainrender/index.html).
Furthermore, an open-access journal article describing BrainRender has been published in eLife, available [here](https://doi.org/10.7554/eLife.65751).
## Installation
From PyPI:
```
pip install brainrender
```
## Quickstart
``` python
import random
import numpy as np
from brainrender import Scene
from brainrender.actors import Points
def get_n_random_points_in_region(region, N):
"""
Gets N random points inside (or on the surface) of a mesh
"""
region_bounds = region.mesh.bounds()
X = np.random.randint(region_bounds[0], region_bounds[1], size=10000)
Y = np.random.randint(region_bounds[2], region_bounds[3], size=10000)
Z = np.random.randint(region_bounds[4], region_bounds[5], size=10000)
pts = [[x, y, z] for x, y, z in zip(X, Y, Z)]
ipts = region.mesh.inside_points(pts).coordinates
return np.vstack(random.choices(ipts, k=N))
# Display the Allen Brain mouse atlas.
scene = Scene(atlas_name="allen_mouse_25um", title="Cells in primary visual cortex")
# Display a brain region
primary_visual = scene.add_brain_region("VISp", alpha=0.2)
# Get a numpy array with (fake) coordinates of some labelled cells
coordinates = get_n_random_points_in_region(primary_visual, 2000)
# Create a Points actor
cells = Points(coordinates)
# Add to scene
scene.add(cells)
# Add label to the brain region
scene.add_label(primary_visual, "Primary visual cortex")
# Display the figure.
scene.render()
```
## Seeking help or contributing
We are always happy to help users of our tools, and welcome any contributions. If you would like to get in contact with us for any reason, please see the [contact page of our website](https://brainglobe.info/contact.html).
## Citing brainrender
If you use brainrender in your scientific work, please cite:
```
Claudi, F., Tyson, A. L., Petrucco, L., Margrie, T.W., Portugues, R., Branco, T. (2021) "Visualizing anatomically registered data with Brainrender" eLife 2021;10:e65751 [doi.org/10.7554/eLife.65751](https://doi.org/10.7554/eLife.65751)
```
BibTeX:
``` bibtex
@article{Claudi2021,
author = {Claudi, Federico and Tyson, Adam L. and Petrucco, Luigi and Margrie, Troy W. and Portugues, Ruben and Branco, Tiago},
doi = {10.7554/eLife.65751},
issn = {2050084X},
journal = {eLife},
pages = {1--16},
pmid = {33739286},
title = {{Visualizing anatomically registered data with brainrender}},
volume = {10},
year = {2021}
}
```