https://github.com/brainglobe/cellfinder-napari
Efficient cell detection in large images using cellfinder in napari
https://github.com/brainglobe/cellfinder-napari
cell-detection cellfinder keras napari napari-plugin object-detection resnet tensorflow
Last synced: 16 days ago
JSON representation
Efficient cell detection in large images using cellfinder in napari
- Host: GitHub
- URL: https://github.com/brainglobe/cellfinder-napari
- Owner: brainglobe
- License: bsd-3-clause
- Archived: true
- Created: 2021-01-22T13:21:20.000Z (over 4 years ago)
- Default Branch: main
- Last Pushed: 2024-01-05T09:13:25.000Z (almost 2 years ago)
- Last Synced: 2025-09-09T19:20:25.638Z (about 1 month ago)
- Topics: cell-detection, cellfinder, keras, napari, napari-plugin, object-detection, resnet, tensorflow
- Language: Python
- Homepage: https://brainglobe.info/documentation/cellfinder/index.html
- Size: 34.3 MB
- Stars: 22
- Watchers: 3
- Forks: 5
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
- awesome-biological-image-analysis - Cellfinder-napari - Efficient cell detection in large images using [cellfinder](https://brainglobe.info/cellfinder) in napari. (Neuroscience)
README
# This package has moved
`cellfinder-napari` has merged with it's backend code and is now available as a [single package called `cellfinder`](https://github.com/brainglobe/cellfinder).
We recommend you uninstall `cellfinder-napari` and instead use the functionality provided in the `cellfinder` package.
These changes are part of our [wider restructuring](https://brainglobe.info/blog/version1/version_1_announcement.html) of the BrainGlobe suite of tools and analysis pipelines, which you can [keep up to date with on our blog](https://brainglobe.info/blog/index.html).
---
# cellfinder-napari
[](https://github.com/napari/cellfinder-napari/raw/master/LICENSE)
[](https://pypi.org/project/cellfinder-napari)
[](https://python.org)
[](https://github.com/brainglobe/cellfinder-napari/actions)
[](https://codecov.io/gh/brainglobe/cellfinder-napari)
[](https://pepy.tech/project/cellfinder-napari)
[](https://pypi.org/project/cellfinder)
[](https://github.com/brainglobe/cellfinder-napari)
[](https://github.com/python/black)
[](https://pycqa.github.io/isort/)
[](https://github.com/pre-commit/pre-commit)
[](https://docs.brainglobe.info/cellfinder/contributing)
[](https://brainglobe.info/documentation/cellfinder/index.html)
[](https://twitter.com/brain_globe)
### Efficient cell detection in large images (e.g. whole mouse brain images)
`cellfinder-napari` is a front-end to [cellfinder-core](https://github.com/brainglobe/cellfinder-core) to allow ease of use within the [napari](https://napari.org/index.html) multidimensional image viewer. For more details on this approach, please see [Tyson, Rousseau & Niedworok et al. (2021)](https://doi.org/10.1371/journal.pcbi.1009074). This algorithm can also be used within the original
[cellfinder](https://github.com/brainglobe/cellfinder) software for
whole-brain microscopy analysis.
`cellfinder-napari`, `cellfinder` and `cellfinder-core` were developed by [Charly Rousseau](https://github.com/crousseau) and [Adam Tyson](https://github.com/adamltyson) in the [Margrie Lab](https://www.sainsburywellcome.org/web/groups/margrie-lab), based on previous work by [Christian Niedworok](https://github.com/cniedwor), generously supported by the [Sainsbury Wellcome Centre](https://www.sainsburywellcome.org/web/).
----
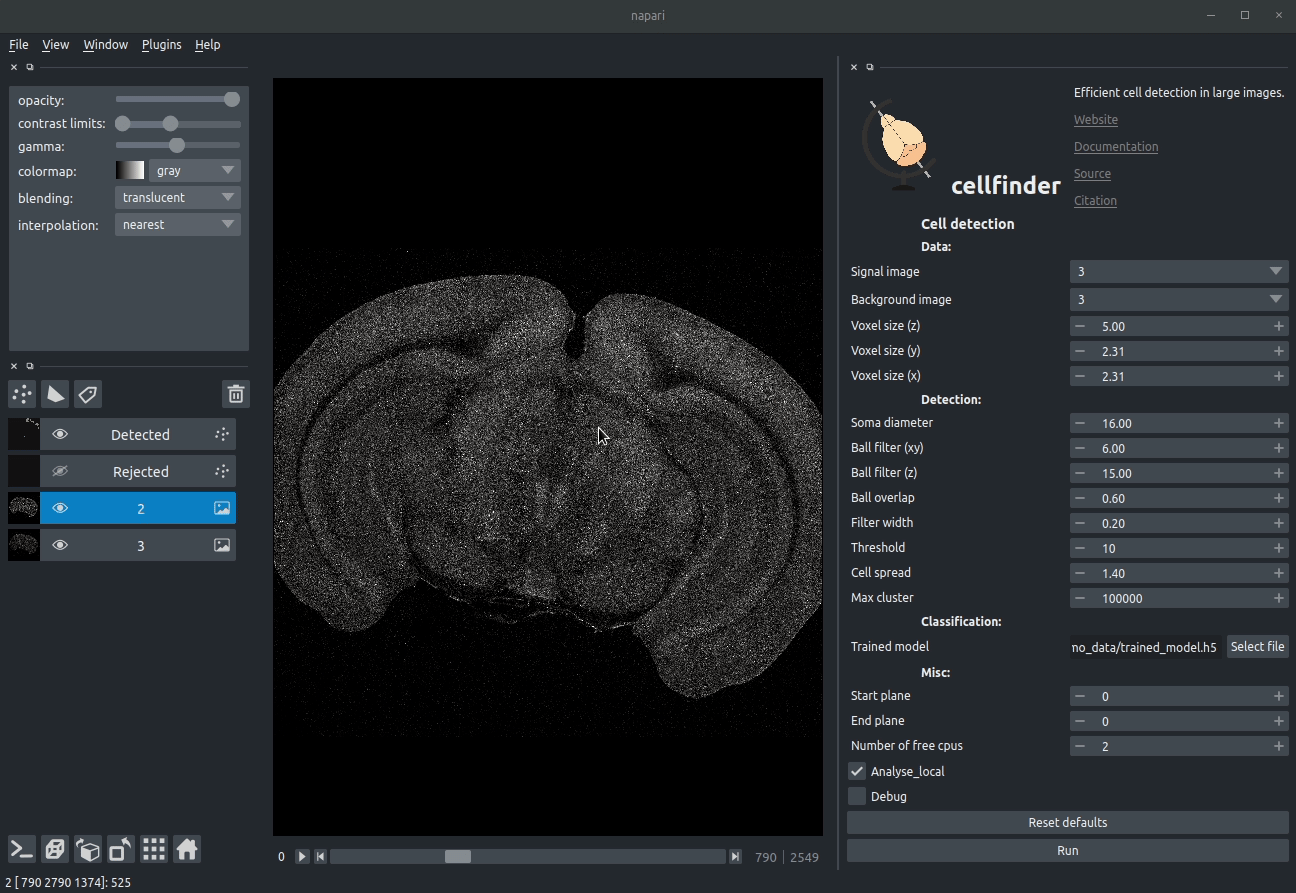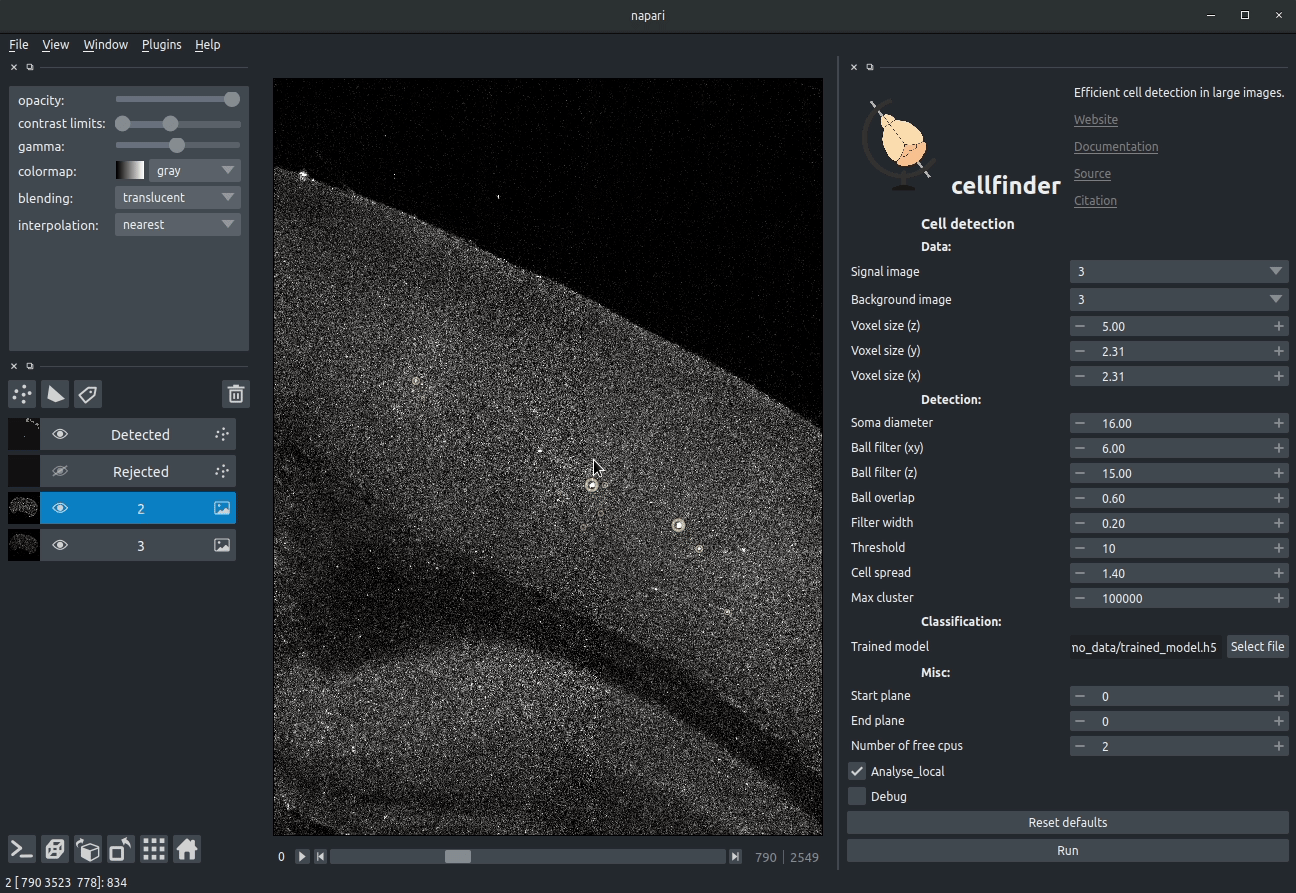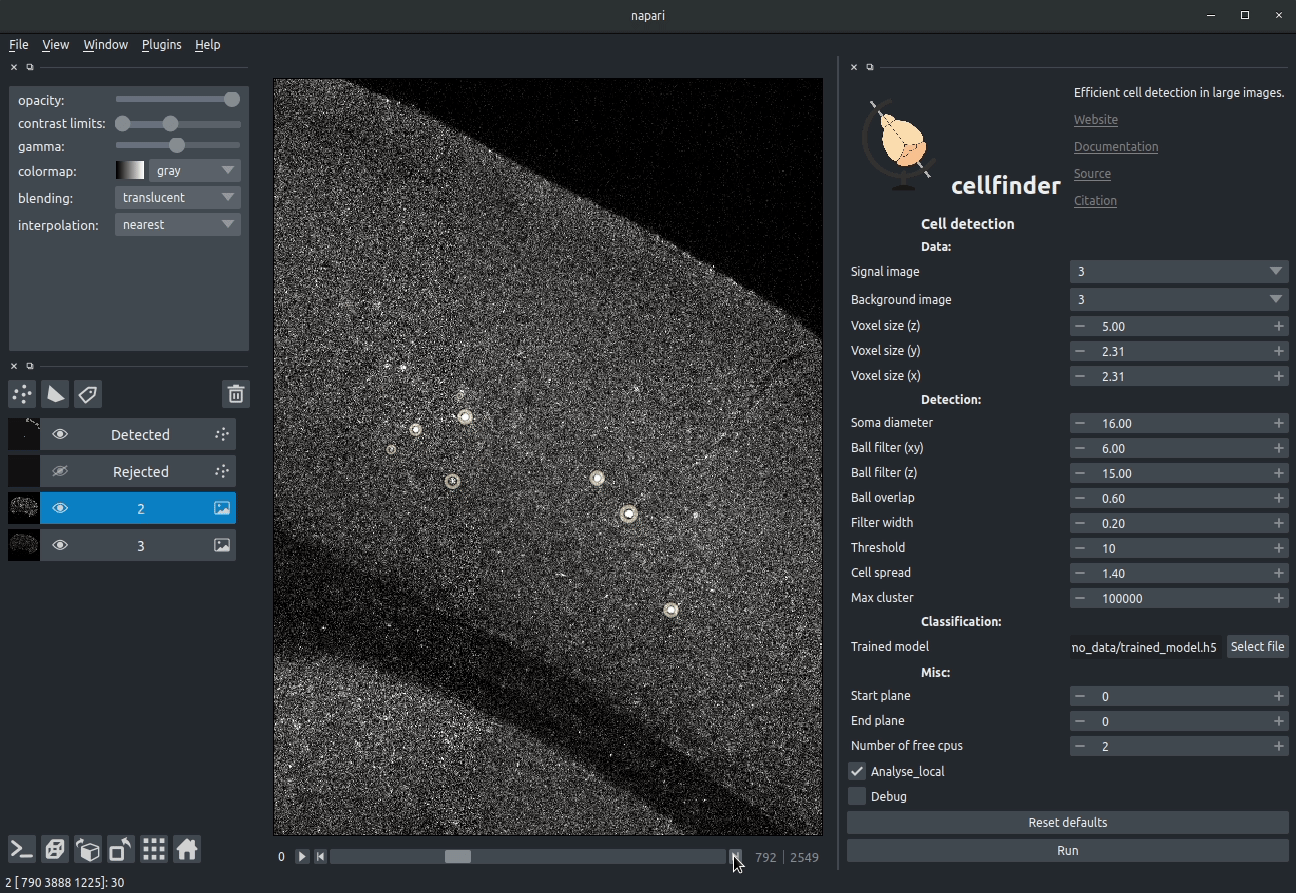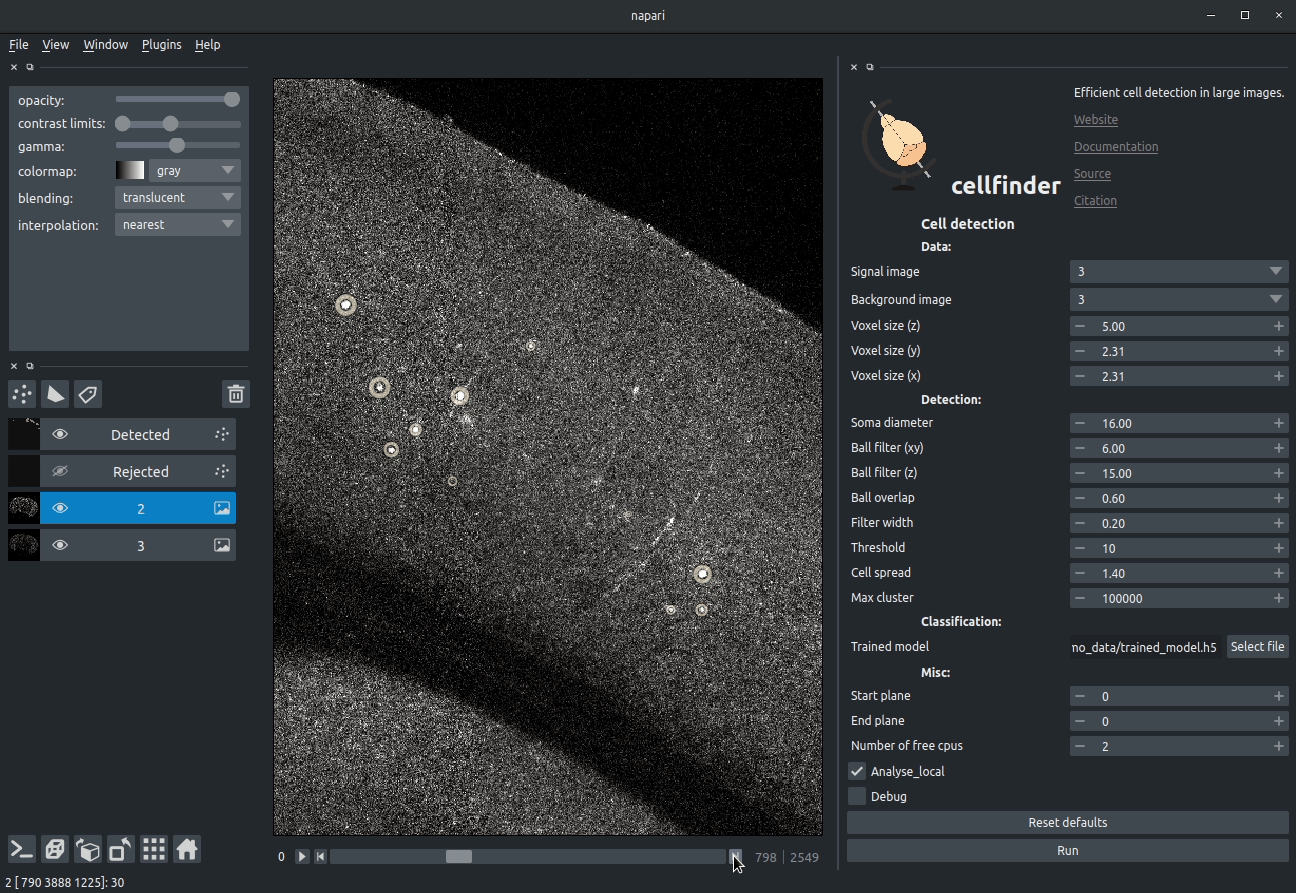
**Visualising detected cells in the cellfinder napari plugin**
----
## Instructions
### Installation
Once you have [installed napari](https://napari.org/index.html#installation).
You can install napari either through the napari plugin installation tool, or
directly from PyPI with:
```bash
pip install cellfinder-napari
```
### Usage
Full documentation can be
found [here](https://brainglobe.info/documentation/cellfinder/index.html).
This software is at a very early stage, and was written with our data in mind.
Over time we hope to support other data types/formats. If you have any
questions or issues, please get in touch [on the forum](https://forum.image.sc/tag/brainglobe) or by
[raising an issue](https://github.com/brainglobe/cellfinder-napari/issues).
---
## Illustration
### Introduction
cellfinder takes a stitched, but otherwise raw dataset with at least
two channels:
* Background channel (i.e. autofluorescence)
* Signal channel, the one with the cells to be detected:

**Raw coronal serial two-photon mouse brain image showing labelled cells**
### Cell candidate detection
Classical image analysis (e.g. filters, thresholding) is used to find
cell-like objects (with false positives):

**Candidate cells (including many artefacts)**
### Cell candidate classification
A deep-learning network (ResNet) is used to classify cell candidates as true
cells or artefacts:

**Cassified cell candidates. Yellow - cells, Blue - artefacts**
## Contributing
Contributions to cellfinder-napari are more than welcome. Please see the [developers guide](https://brainglobe.info/developers/index.html).
## Citing cellfinder
If you find this plugin useful, and use it in your research, please cite the paper outlining the cell detection algorithm:
> Tyson, A. L., Rousseau, C. V., Niedworok, C. J., Keshavarzi, S., Tsitoura, C., Cossell, L., Strom, M. and Margrie, T. W. (2021) “A deep learning algorithm for 3D cell detection in whole mouse brain image datasets’ PLOS Computational Biology, 17(5), e1009074
[https://doi.org/10.1371/journal.pcbi.1009074](https://doi.org/10.1371/journal.pcbi.1009074)
**If you use this, or any other tools in the brainglobe suite, please
[let us know](mailto:code@adamltyson.com?subject=cellfinder-napari), and
we'd be happy to promote your paper/talk etc.**