https://github.com/brj0/mepylome
Python toolkit for parsing, processing, and analysis of Illumina methylation array IDAT files
https://github.com/brj0/mepylome
cnv conumee epigenetics illumina methylation methylation-analysis methylation-extractor methylation-microarrays methylation-plot minfi python tumor-classification umap
Last synced: 3 days ago
JSON representation
Python toolkit for parsing, processing, and analysis of Illumina methylation array IDAT files
- Host: GitHub
- URL: https://github.com/brj0/mepylome
- Owner: brj0
- License: mit
- Created: 2024-02-08T07:34:27.000Z (over 1 year ago)
- Default Branch: main
- Last Pushed: 2025-09-23T05:34:59.000Z (16 days ago)
- Last Synced: 2025-09-23T07:19:54.879Z (16 days ago)
- Topics: cnv, conumee, epigenetics, illumina, methylation, methylation-analysis, methylation-extractor, methylation-microarrays, methylation-plot, minfi, python, tumor-classification, umap
- Language: Python
- Homepage: https://mepylome.readthedocs.io
- Size: 14.4 MB
- Stars: 9
- Watchers: 3
- Forks: 1
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README

[](https://badge.fury.io/py/mepylome)
[](https://opensource.org/licenses/MIT)

[](https://mepylome.readthedocs.io/en/latest/?badge=latest)
[](https://colab.research.google.com/github/brj0/mepylome/blob/main/examples/tutorial.ipynb)
-----------------
# Mepylome: Methylation Array Analysis Toolkit
Mepylome is an efficient Python toolkit tailored for parsing, processing, and
analyzing methylation array IDAT files. Serving as a versatile library,
Mepylome supports a wide range of methylation analysis tasks. It also includes
an interactive GUI that enables users to generate UMAP plots and CNV plots
(Copy Number Variation) directly from collections of IDAT files.
## Features
- Parsing of IDAT files
- Extraction of methylation signals
- Calculation of Copy Number Variations (CNV) with visualization using
[plotly](https://github.com/plotly/plotly.py).
- Support for the following Illumina array types: 450k, EPIC, EPICv2, MSA48
- Significantly faster compared to [minfi](https://github.com/hansenlab/minfi)
and [conumee2](https://github.com/hovestadtlab/conumee2).
- Methylation analysis tool with a graphical browser interface for UMAP
analysis, CNV plots and supervised classification
- Can be run from the command line with minimal setup or customized through a
Python script
- CN-summary plots
## Documentation
The mepylome documentation, including installation instructions, tutorial and API, is available at
## Usage
### Methylation extraction and copy number variation plots
```python
from pathlib import Path
from mepylome import CNV, MethylData
# Sample
analysis_dir = Path("/path/to/idat/directory")
sample_file = analysis_dir / "200925700125_R07C01"
# CNV neutral reference files
reference_dir = Path("/path/to/reference/directory")
# Get methylation data
sample_methyl = MethylData(file=sample_file)
reference_methyl = MethylData(file=reference_dir)
# Beta value
betas = sample_methyl.betas
# Print overview of processed data
print(sample_methyl)
# CNV anylsis
cnv = CNV.set_all(sample_methyl, reference_methyl)
# Visualize CNV in the browser
cnv.plot()
```
### Methylation analysis: Command-line interface and GUI
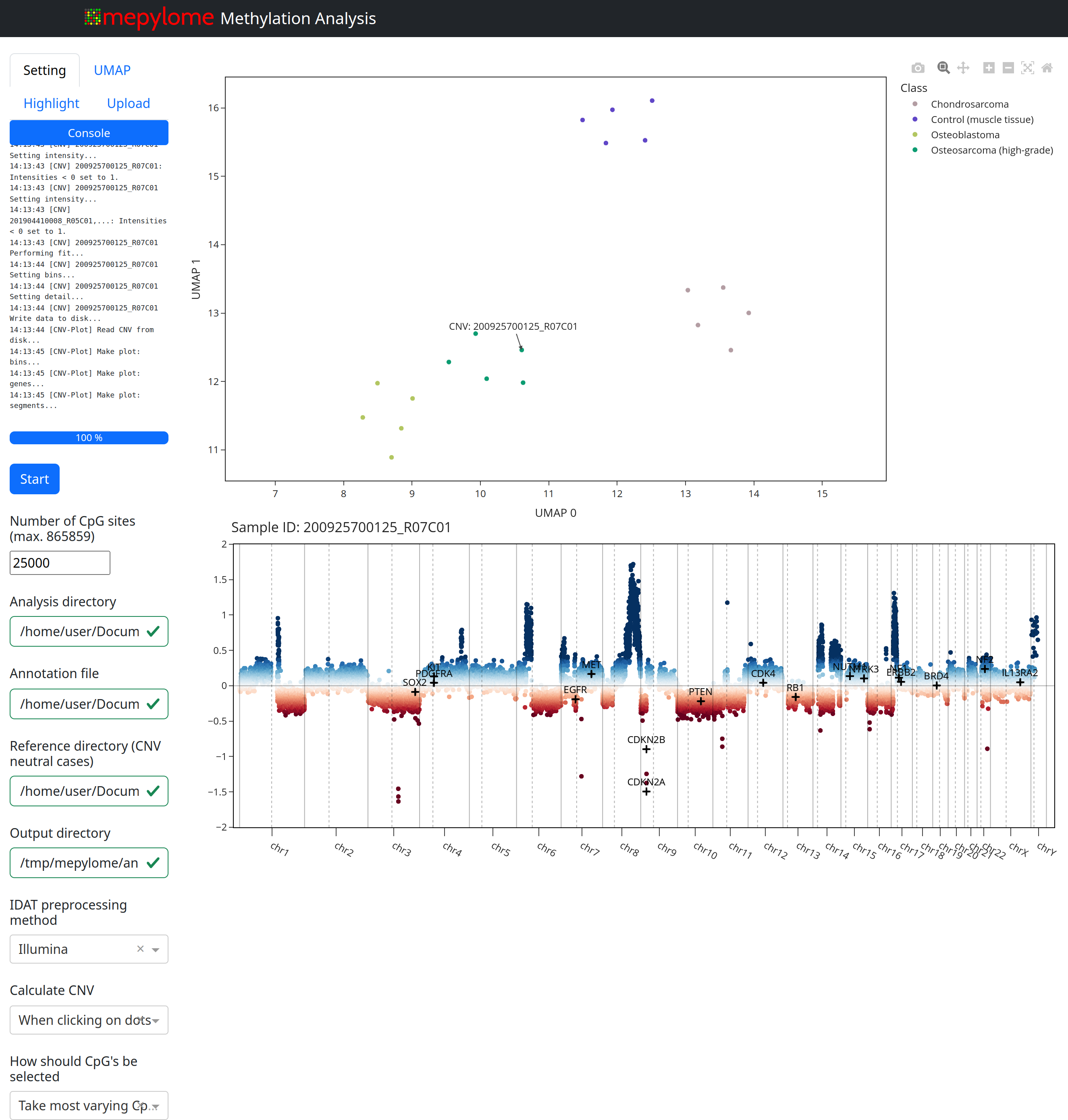
#### Basic usage:
Mepylome provides a command-line interface for launching a GUI and performing
methylation analysis. Ensure you have an analysis directory, a CNV reference
directory, and an annotation file (located within the analysis directory). Use
the following command to initiate the analysis:
```sh
mepylome --analysis_dir /path/to/idats --reference_dir /path/to/ref
```
If you want to perform a **quick test**, use:
```sh
mepylome --tutorial
```
This command downloads sample IDAT files and provides a demonstration of the
package's functionality.
You can try the tutorial directly in **Google Colab**-without downloading or
installing anything-by clicking the link below. Please note that GUI support is
limited in Colab.
[](https://colab.research.google.com/github/brj0/mepylome/blob/main/examples/tutorial.ipynb)
See for details.
## C++ parser
Mepylome also includes a C++ parser. See for
details.
## Contributing
Contributions are welcome! If you have any bug reports, feature requests, or suggestions, please open an issue or submit a pull request.
## License
This project is licensed under the [MIT license](LICENSE).
## Acknowledgements
Mepylome is strongly influenced by [minfi](https://github.com/hansenlab/minfi) and [conumee2](https://github.com/hovestadtlab/conumee2). Some functionalities, such as the manifest handler and parser, are adapted from [methylprep](https://github.com/FoxoTech/methylprep).