https://github.com/computational-cell-analytics/micro-sam
Segment Anything for Microscopy
https://github.com/computational-cell-analytics/micro-sam
cell-segmentation microscopy-images mitochondria-segmentation napari nuclei-segmentation segment-anything segmentation
Last synced: 10 months ago
JSON representation
Segment Anything for Microscopy
- Host: GitHub
- URL: https://github.com/computational-cell-analytics/micro-sam
- Owner: computational-cell-analytics
- License: mit
- Created: 2023-05-03T11:36:51.000Z (almost 3 years ago)
- Default Branch: master
- Last Pushed: 2025-05-04T17:52:57.000Z (10 months ago)
- Last Synced: 2025-05-04T18:33:56.568Z (10 months ago)
- Topics: cell-segmentation, microscopy-images, mitochondria-segmentation, napari, nuclei-segmentation, segment-anything, segmentation
- Language: Jupyter Notebook
- Homepage: https://computational-cell-analytics.github.io/micro-sam/
- Size: 51.7 MB
- Stars: 534
- Watchers: 9
- Forks: 67
- Open Issues: 74
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
- awesome-biological-image-analysis - MicroSAM - Tools for segmentation and tracking in microscopy build on top of SegmentAnything. Segment and track objects in microscopy images interactively. (Image processing and segmentation)
README
[](https://computational-cell-analytics.github.io/micro-sam/)
[](https://anaconda.org/conda-forge/micro_sam)
[](https://codecov.io/gh/computational-cell-analytics/micro-sam)
[](https://doi.org/10.5281/zenodo.7919746)
# Segment Anything for Microscopy
Tools for segmentation and tracking in microscopy build on top of [Segment Anything](https://segment-anything.com/).
Segment and track objects in microscopy images interactively with a few clicks!
We implement napari applications for:
- interactive 2d segmentation (Left: interactive cell segmentation)
- interactive 3d segmentation (Middle: interactive mitochondria segmentation in EM)
- interactive tracking of 2d image data (Right: interactive cell tracking)
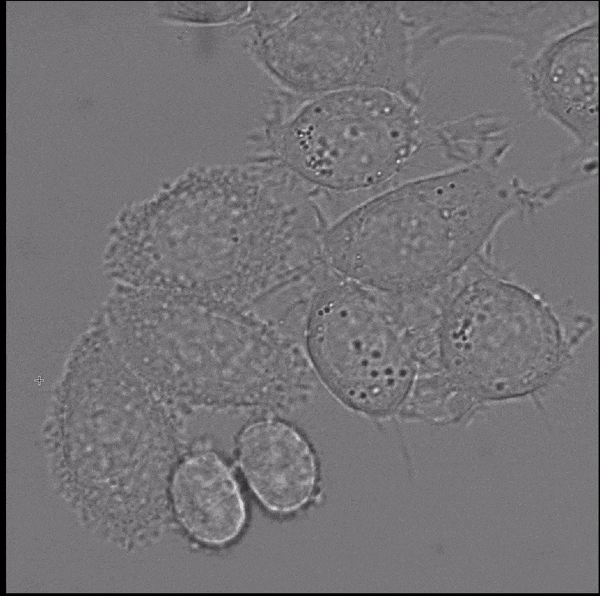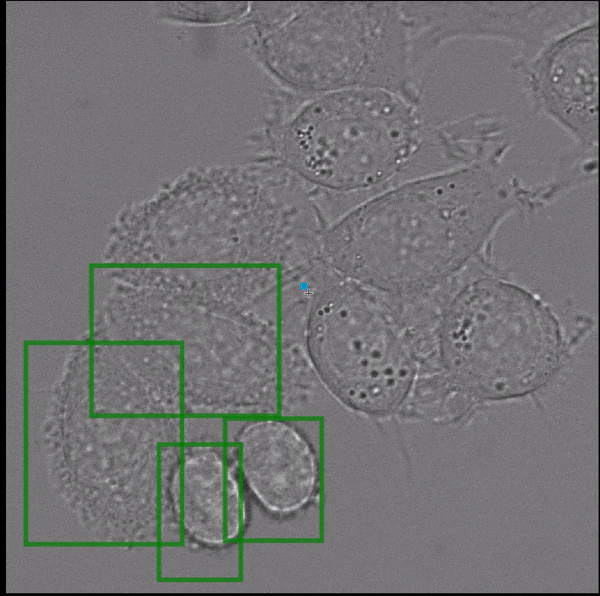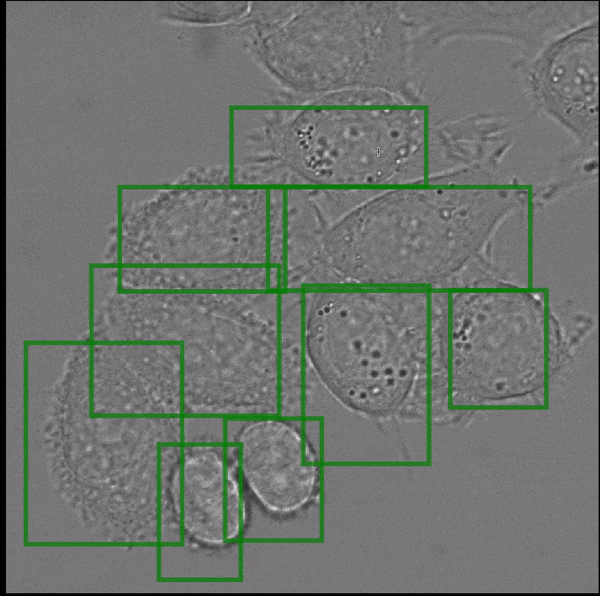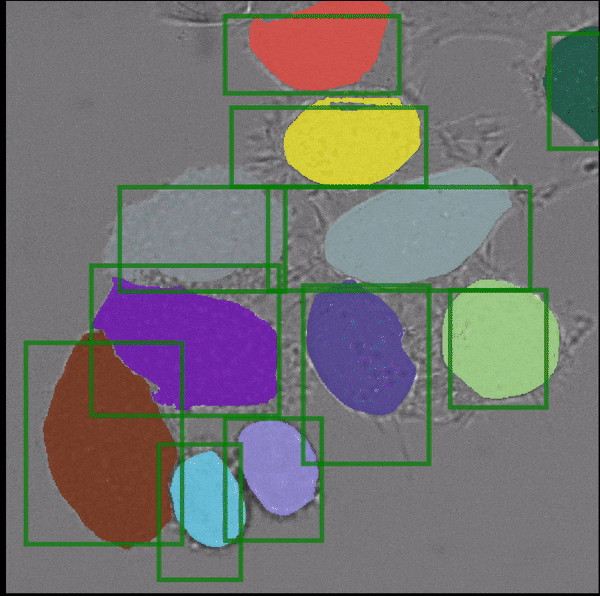
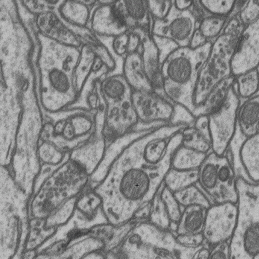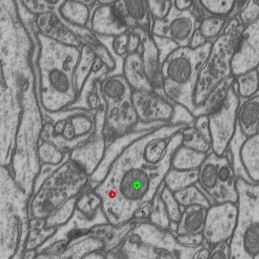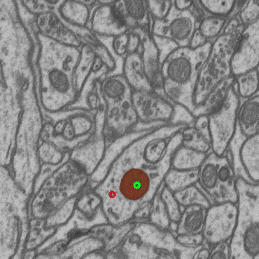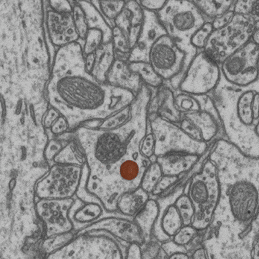
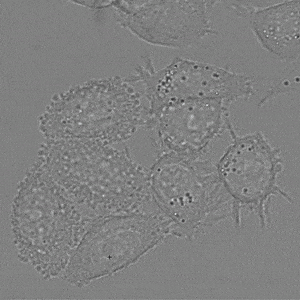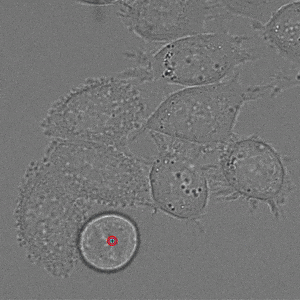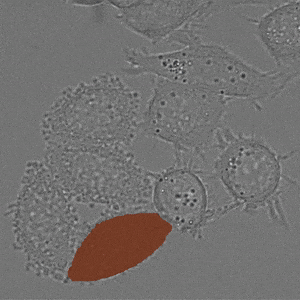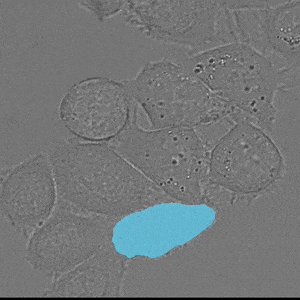
If you run into any problems or have questions regarding our tool please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
## Installation and Usage
Please check [the documentation](https://computational-cell-analytics.github.io/micro-sam/) for details on how to install and use `micro_sam`. You can also watch [the quickstart video](https://youtu.be/gcv0fa84mCc) or [all video tutorials](https://youtube.com/playlist?list=PLwYZXQJ3f36GQPpKCrSbHjGiH39X4XjSO&si=qNbB8IFXqAX33r_Z).
## Contributing
We welcome new contributions!
If you are interested in contributing to micro-sam, please see the [contributing guide](https://computational-cell-analytics.github.io/micro-sam/micro_sam.html#contribution-guide). The first step is to [discuss your idea in a new issue](https://github.com/computational-cell-analytics/micro-sam/issues/new) with the current developers.
## Citation
If you are using this repository in your research please cite
- our [preprint](https://doi.org/10.1101/2023.08.21.554208)
- and the original [Segment Anything publication](https://arxiv.org/abs/2304.02643).
- If you use a `vit-tiny` models please also cite [Mobile SAM](https://arxiv.org/abs/2306.14289).
## Related Projects
There are a few other napari plugins build around Segment Anything:
- https://github.com/MIC-DKFZ/napari-sam (2d and 3d support)
- https://github.com/royerlab/napari-segment-anything (only 2d support)
- https://github.com/hiroalchem/napari-SAM4IS
Compared to these we support more applications (2d, 3d and tracking), and provide finetuning methods and finetuned models for microscopy data.
[WebKnossos](https://webknossos.org/) also offers integration of SegmentAnything for interactive segmentation.
## Release Overview
**New in version 1.0.1**
Use stable URL for model downloads and fix issues in state precomputation for automatic segmentation.
**New in version 1.0.0**
This release mainly fixes issues with the previous release and marks the napari user interface as stable.
**New in version 0.5.0**
This version includes a lot of new functionality and improvements. The most important changes are:
- Re-implementation of the annotation tools. The tools are now implemented as napari plugin.
- Using our improved functionality for automatic instance segmentation in the annotation tools, including automatic segmentation for 3D data.
- New widgets to use the finetuning and image series annotation functionality from napari.
- Improved finetuned models for light microscopy and electron microscopy data that are available via bioimage.io.
**New in version 0.4.1**
- Bugfix for the image series annotator. Before the automatic segmentation did not work correctly.
**New in version 0.4.0**
- Significantly improved model finetuning
- Update the finetuned models for microscopy, see [details in the doc](https://computational-cell-analytics.github.io/micro-sam/micro_sam.html#finetuned-models)
- Training decoder for direct instance segmentation (not available via the GUI yet)
- Refactored model download functionality using [pooch](https://pypi.org/project/pooch/)
**New in version 0.3.0**
- Support for ellipse and polygon prompts
- Support for automatic segmentation in 3d
- Training refactoring and speed-up of fine-tuning
**New in version 0.2.1 and 0.2.2**
- Several bugfixes for the newly introduced functionality in 0.2.0.
**New in version 0.2.0**
- Functionality for training / finetuning and evaluation of Segment Anything Models
- Full support for our finetuned segment anything models
- Improvements of the automated instance segmentation functionality in the 2d annotator
- And several other small improvements
**New in version 0.1.1**
- Fine-tuned segment anything models for microscopy (experimental)
- Simplified instance segmentation menu
- Menu for clearing annotations
**New in version 0.1.0**
- We support tiling in all annotators to enable processing large images.
- Implement new automatic instance segmentation functionality:
- That is faster.
- Enables interactive update of parameters.
- And also works for large images by making use of tiled embeddings.
- Implement the `image_series_annotator` for processing many images in a row.
- Use the data hash in pre-computed embeddings to warn if the input data changes.
- Create a simple GUI to select which annotator to start.
- And made many other small improvements and fixed bugs.
**New in version 0.0.2**
- We have added support for bounding box prompts, which provide better segmentation results than points in many cases.
- Interactive tracking now uses a better heuristic to propagate masks across time, leading to better automatic tracking results.
- And have fixed several small bugs.