https://github.com/cytomining/cytodataframe
An in-memory data analysis format for single-cell profiles alongside their corresponding images and segmentation masks.
https://github.com/cytomining/cytodataframe
dataframes image-analysis image-based-profiling single-cell
Last synced: 7 months ago
JSON representation
An in-memory data analysis format for single-cell profiles alongside their corresponding images and segmentation masks.
- Host: GitHub
- URL: https://github.com/cytomining/cytodataframe
- Owner: cytomining
- License: bsd-3-clause
- Created: 2024-10-22T22:47:10.000Z (about 1 year ago)
- Default Branch: main
- Last Pushed: 2025-06-11T16:50:05.000Z (7 months ago)
- Last Synced: 2025-06-11T18:12:10.604Z (7 months ago)
- Topics: dataframes, image-analysis, image-based-profiling, single-cell
- Language: Python
- Homepage: https://cytomining.github.io/CytoDataFrame
- Size: 69.3 MB
- Stars: 2
- Watchers: 1
- Forks: 2
- Open Issues: 16
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
- Code of conduct: CODE_OF_CONDUCT.md
- Citation: CITATION.cff
Awesome Lists containing this project
README
# CytoDataFrame
[](https://pypi.org/project/CytoDataFrame/)
[](https://github.com/cytomining/CytoDataFrame/actions/workflows/run-tests.yml?query=branch%3Amain)

[](https://github.com/astral-sh/ruff)
[](https://python-poetry.org/)
[](https://doi.org/10.5281/zenodo.14797074)
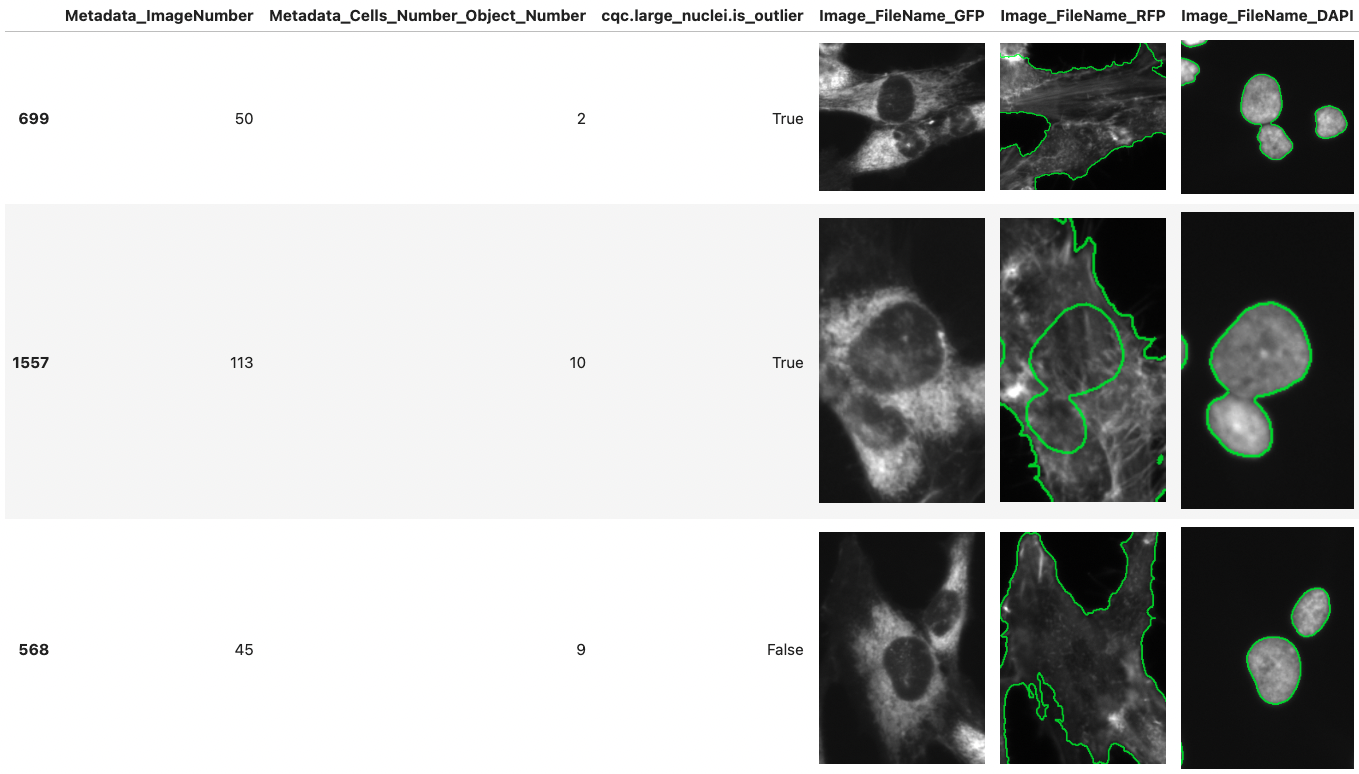
_CytoDataFrame extends Pandas functionality to help display single-cell profile data alongside related images._
CytoDataFrame is an advanced in-memory data analysis format designed for single-cell profiling, integrating not only the data profiles but also their corresponding microscopy images and segmentation masks.
Traditional single-cell profiling often excludes the associated images from analysis, limiting the scope of research.
CytoDataFrame bridges this gap, offering a purpose-built solution for comprehensive analysis that incorporates both the data and images, empowering more detailed and visual insights in single-cell research.
CytoDataFrame is best suited for work within Jupyter notebooks.
With CytoDataFrame you can:
- View image objects alongside their feature data using a Pandas DataFrame-like interface.
- Highlight image objects using mask or outline files to understand their segmentation.
- Adjust image displays on-the-fly using interactive slider widgets.
📓 ___Want to see CytoDataFrame in action?___ Check out our [example notebook](docs/src/examples/cytodataframe_at_a_glance.ipynb) for a quick tour of its key features.
> ✨ CytoDataFrame development began within **[coSMicQC](https://github.com/cytomining/coSMicQC)** - a single-cell profile quality control package.
> Please check out our work there as well!
## Installation
Install CytoDataFrame from source using the following:
```shell
# install from pypi
pip install cytodataframe
# or install directly from source
pip install git+https://github.com/cytomining/CytoDataFrame.git
```
## Contributing, Development, and Testing
Please see our [contributing](https://cytomining.github.io/CytoDataFrame/main/contributing) documentation for more details on contributions, development, and testing.
## References
- [coSMicQC](https://github.com/cytomining/coSMicQC)
- [pycytominer](https://github.com/cytomining/pycytominer)
- [CellProfiler](https://github.com/CellProfiler/CellProfiler)
- [CytoTable](https://github.com/cytomining/CytoTable)