https://github.com/deepmodeling/dpti
A Python Package to Automate Thermodynamic Integration Calculations for Free Energy
https://github.com/deepmodeling/dpti
atomistic-simulations free-energy molecular-dynamics thermodynamic-integration workflow
Last synced: 5 months ago
JSON representation
A Python Package to Automate Thermodynamic Integration Calculations for Free Energy
- Host: GitHub
- URL: https://github.com/deepmodeling/dpti
- Owner: deepmodeling
- License: lgpl-3.0
- Created: 2021-04-04T13:57:37.000Z (almost 5 years ago)
- Default Branch: devel
- Last Pushed: 2025-10-06T21:20:07.000Z (5 months ago)
- Last Synced: 2025-10-06T23:23:11.149Z (5 months ago)
- Topics: atomistic-simulations, free-energy, molecular-dynamics, thermodynamic-integration, workflow
- Language: Python
- Homepage: https://docs.deepmodeling.com/projects/dpti
- Size: 779 KB
- Stars: 30
- Watchers: 5
- Forks: 20
- Open Issues: 13
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
- deepmodeling-projects - GitHub - 90% open · ⏱️ 15.03.2025): (Others)
README
deepmodeling, deepmd, dpti, free energy, phase diagram
---
# 🏞brief Introduction
**dpti** (deep potential thermodynamic integration) is a python package for calculating free energy, doing thermodynamic integration and figuring out pressure-temperature phase diagram for materials with molecular dynamics (MD) simulation methods.
The user will get Gibbs (Helmholtz) free energy of a system at different temperature and pressure conditions. With these free energy results, the user could determine the phase transition points and coexistence curve on the pressure-volume phase diagram.
useful docs:
github README.md: [https://github.com/deepmodeling/dpti/README.md](https://github.com/deepmodeling/dpti/README.md)
some discussion notes: [https://www.yuque.com/dpti/manual/ilzmlb](https://www.yuque.com/dpti/manual/ilzmlb)
intruction to free energy calculation:
[https://nb.bohrium.dp.tech/detail/18465833825](https://nb.bohrium.dp.tech/detail/18465833825)
## for water and ice
see: [PRL:Phase Diagram of a Deep Potential Water Model](https://journals.aps.org/prl/abstract/10.1103/PhysRevLett.126.236001)
Phase diagram of water. DP model (red solid lines) and experiment (gray solid lines) for $T<420K$. Black letters indicate phases that are stable in the experiment and model. The original figure is in article。

## for metal Tin(Sn)
We could use dpti to calculate out the Press-Volume phase diagram of metals.
The picture below shows the metal Sn phase diagram results calculated by one of the authors.
The left subgraph shows the experiment phase diagram results.(see:[https://aip.scitation.org/doi/10.1063/1.4872458](https://aip.scitation.org/doi/10.1063/1.4872458))
The middle subgraph shows the DP phase diagram based on SCAN functional DFT calculation results.
The right subgraph shows the DP phase diagram base on PBE functional DFT calculation results.

# 🦴software introduction
At first, dpti is a collection of python scripts to generate LAMMPS input scripts and to anaylze results from LAMMPS logs.
In dpti, there are many MD simulations tasks and scripts need to be run sequentially or concurrently. Before and after these MD simulation tasks, we may run a lot of MD scirpts to prepare the input files or analyze the logs to extract the useful data.
Then the dpti developers use apache-airflow to resolve the MD tasks dependencies and managing running tasks.
# Software Usage:
>the examples dir `examples/` in source code contains the essential files and jsons.
## for CLI tools:
The following scripts can be used by Python CLI to generate essential scripts for LAMMPS simulation.
the CLI entry:
```
# after installation: pip install .
dpti --help
```
### Equi(npt and nvt simulation)
The following scripts are used to generate essential tools.
```
cd exampls/equi/
dpti equi --help`
dpti equi gen npt.json
dpti equi gen nvt.json
```
The dir `new_job/` contains the simulation files
### HTI
This is an example for HTI three-step simulations.
```
cd examples/hti/
dpti hti --help
dpti hti gen hti.json -s three-step
```
### TI
For temperature 200K, in order to generate integration path pressure from 0 to 10000 of interval 500.
in `ti.p.json`, we writes
```json
"temp":200,
"pres_seq":[
"0:10000:500",
"10000"
]
```
```
cd examples/ti/
dpti ti gen ti.t.json
```
In order to generate TI path changing temperature, we use
```
dpti ti gen ti.p.json
```
### GDI
An example for finding coexisting line between Sn `beta` and `alpha` phase.
by `gdidata.json`:
starting point is 1GPa,270K. (calculated by HTI method)
We want to extend to 1.35 GPa.
```
cd examples/gdi/
dpti gdi pb.json machine.json -g gdidata.json
```
## For airflow workflow:
Sometimes, we need to do **high-throughput** calculations(which means we need to calculate a series of temperature, pressure points for multiple phases).
It would be a great burden for users to execute these tasks manually and monitor the tasks' execution.
We provide the workflow tools based on apache-airflow workflow framework.
>we refer this docs [airflow official docs](https://airflow.apache.org/docs/apache-airflow/stable/index.html) for more instructions.
### TI_Workflow
We implement a workflow de
implemented at `workflow/DpFreeEnergy.py`
example dir and json:
```
cd examples/
cat examples/FreeEnergy.json
```
Requirement: setup apache-airflow or use the docker version dpti
```
docker run --name dpti -p 9999:8080 -it deepmodeling/dpti:latest /bin/bash
docker exec -it dpti /bin/bash
```
Then we provide a basic example for apache-airflow usage
```
# pwd at /home/airflow/
cd dpti/examples/
airflow dags trigger TI_taskflow --conf $(printf "%s" $(cat FreeEnergy.json))
```
**Input:** Lammps structure file.
**Output:** free energy values at given temperature and pressure.
**Parameters:** Given temperature(or the range), Given pressure(or therange), force field,Lammps simulation ensemble and etc.
we implement a workflow called TI_taskflow:
It includes these steps:
1. npt simulation to get lattice constant.
2. nvt simulation.
3. HTI: free energy at given temperature and pressure
4. TI: free energy values at the given range of temperature/pressure.
### website to manage and monitor these jobs.
``
## local CLI tools installtion
```
# usually create a new python environment
# conda create --name dpti
# conda activate dpti
cd dpti/
pip install .
# use this command to check installation
dpti --help
```
## docker image:
```
docker pull deepmodeling/dpti
```
The useful files and command see [this file](docker/README.md
## Manually installation
>the [Dockerfile](docker/Dockerfile) at `docker/` dir may be helpful for mannually installation.
dpti use apache-airflow as workflow framework, and dpdispatcher to interact with the HPC systems (slurm or PBS).
airflow use realation database (PostgreSQL, MySQL or Sqlite) as backend to store the metadata, DAGs definetion and nodes state etc.
### install dpti and dpdispatcher.
git clone the following packages and install.
[https://github.com/deepmodeling/dpti](https://github.com/deepmodeling/dpti)
```bash
cd dpti/
pip install .
```
### install postgresql backend
apahche-airflow require a database backend.Here we refer to this doc [postgresql offical docs for download](https://www.postgresql.org/download/)
and use this command
```
psql -h
```
### configure apache-airflow.
airflow user manual: [https://airflow.apache.org/docs/apache-airflow/stable/index.html](https://airflow.apache.org/docs/apache-airflow/stable/index.html)
```bash
# airflow will create at ~/airflow
airflow -h
cd ~/airflow
# usually the configuration file location
# we refer this doc for further information
# https://airflow.apache.org/docs/apache-airflow/stable/configurations-ref.html
vi ~/airflow/airflow.cfg
# airflow will initialize datebase with sqlite
airflow db init
# create a user
airflow users create \
--username airflow \
--firstname Peter \
--lastname Parker \
--role Admin \
--email spiderman@superhero.org
# you will be requested to enter the password here.
# start airflow's webserver to manage your workflow use "-D" option to daemon it
airflow webserver --port 8080 --hostname 127.0.0.1
# start airflwo scheduler
airflow scheduler
# if ariflow web server start at the personal computer,
# you could go to http://localhost:8080/ to view it
# if airflow runs on remote server
# you could use ssh to conntect to server
# ssh -CqTnN -L localhost:8080:localhost:8080 someusername@39.xx.84.xx
```
## with docker
```docker pull deepmodeling/dpti```
further information(useful command and files, examples):see [docker README.md](docker/README.md)
## manually
```bash
# copy dpti'workflow file
cp /path-to-dpti/workflow/DpFreeEnergy.py ~/airflow/dags/
# create a workdir and copy example files
cp /path-to-dpti/examples/*json /path-to-a-work-dir/
# start our airflow job
cd /path-to-a-work-dir/
cat ./airflow.sh
airflow dags trigger TI_taskflow --conf $(printf "%s" $(cat FreeEnergy.json))
```
##
## 🕹install postgresql database
Airflow use relation database as backend. And PostgreSQL is widely used in airflow community.
### install database
airflow's introduction on how to set up database backend: [apache-airflow:set up database](https://airflow.apache.org/docs/apache-airflow/stable/howto/set-up-database.html)
```bash
# install apache-airflow postgresql module
pip install apache-airflow-providers-postgres
# install postgresql
yum install postgresql
# enable postgresql service
systemctl start postgresql
# enter posgresql
psql
```
### create database and database user
```sql
CREATE DATABASE airflow_db1;
CREATE USER airflow_user1 WITH PASSWORD 'airflow_user1';
GRANT ALL PRIVILEGES ON DATABASE airflow_db1 TO airflow_user1;
```
###
### configure airflow configure file to connect database
configure ~/airflow/airflow.cfg
```
# change the following item with the sql above
# sql_alchemy_conn = sqlite:////home/fengbo/airflow/airflow.db
# sql_alchemy_conn = postgres://airflow:airflow@localhost:5432/airflow
sql_alchemy_conn = postgresql+psycopg2://:@:/
```
### configure apache-airflow
reset db and webserver scheduler
```
# reset db
airflow db init
# -D flag represent daemonize
airflow webserver # -p 8080 -D
airflow scheduler # -D
```
### airflow webserver
If things work well, we could type the ip and prot in the web Browser and use the web service to monitor and manage the tasks operated by apache-airflow.
We refer to this doc [apache-airflow webserver guide](https://airflow.apache.org/docs/apache-airflow/stable/security/webserver.html) for further information.
after login(usually with default username and password:airflow, airflow)
[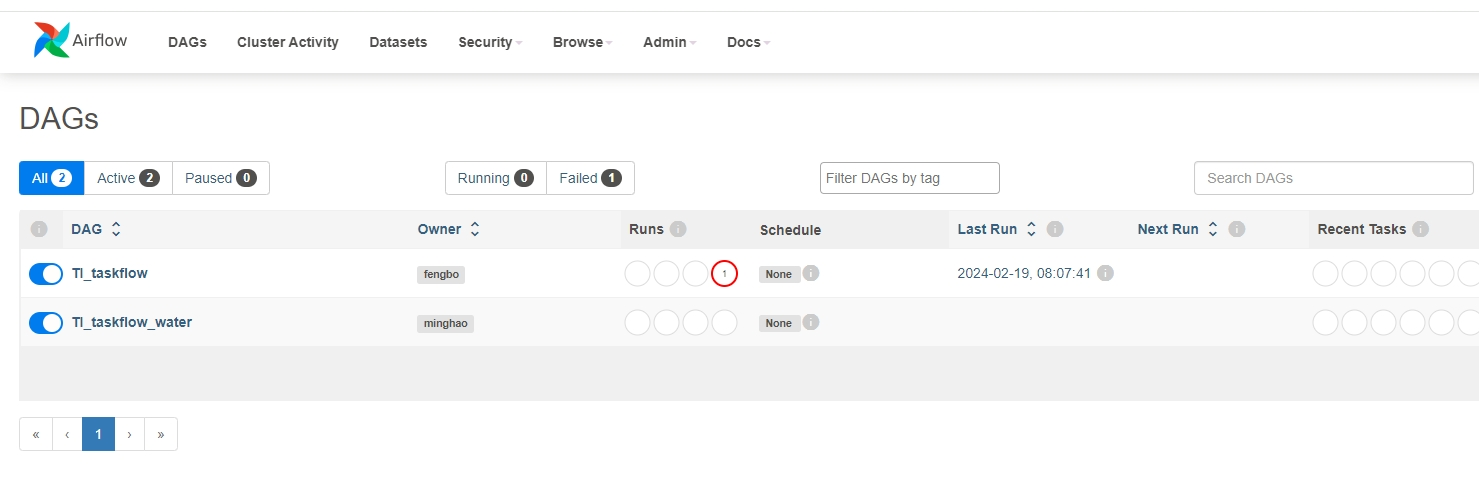](https://imgse.com/i/pFYmlWj)
### ssh to use the webserver
Sometime, apache-airflow runs on remote machine and user can `ssh` to contect to the cloud server by command like ` ssh -L localhost:8080:localhost:8080 user1@67.xxx.xxx.25` and visit [http://localhost:8080/](http://localhost:8080/) to monitor the free energy calculation tasks process.
### apache-airflow: further instruction
The backend of this software is based on the software `airflow`. The following command can start the calculation.
The first command is used for calculate the free energy of solid.
The second command is used for calculate the free energy of liquid.
```bash
airflow trigger_dag HTI_taskflow --conf $(printf "%s" $(cat FreeEnergy.json))
airflow trigger_dag TI_taskflow --conf $(printf "%s" $(cat FreeEnergy.meam.json))
```
#### FreeEnergy.json
We usually want to calculate the free energy of a metal at a specific pressure or temperature. And the crystal structure of the metal can be various. For example, we want to calculate the free energy of metal Sn of bcc structure at 200 K and 50000 bar (5GPa). In order to caculate the per atom free energy of metal Sn. First, We must prepare a configuration file named bcc.lmp and modify the [FreeEnergy.json](#ULX0o) or [FreeEnergyLiquid.json](#WuLBQ) and modify the key-value pair like "structure": "bcc", "target_temp": 200, "target_press" : 50000. And decide whether to integrate along the t(temperature) path or along the p(pressure) path . Modify the "path" key-value pair for this. The key-value pair "ensemble" for lammps MD simulation. Usually the ensemble shoule be consistent with the crystal intrinsic structure. That means we should set "npt-iso" for structure "bcc" to keep the simulation box changes simultaneously in x, y, z directions.
#### ti-path json
Modify the ti.t.json or ti.p.json, and change the key-value pair "temps" or "press" . For ti.t.json, the tar_temp of FreeEnergy.json must be in the list which the key-value pair "temps" of ti.t.json represents. And similarly for ti.p.json, the tar_press of FreeEnergy.json must be in the list which the key-value pair "temps" of ti.t.json represents.
#### workflow
1. Use the command `airflow trigger_dag` mentioned above. This command will start a [airflow dag](https://airflow.apache.org/docs/apache-airflow/stable/concepts.html).This dag is wrote and maintained by the dpti software developer. It is used to make the calculation to be done more autocally . The user could monitor the task state and calculation procedure at [a website](#2aabcbd6). The user can also rerun, restart, delete the whole calculation or some part of the calculations.
2. Wait until the calculation finish. Usually the whole procedure continues for about 6 to 10 hours. The calculations will be done autocally.
3. Find the results in [Results Show](#2aabcbd6) part. The user could use the tables and data of it and plot the curve.
# 📌Calculation Results and files
For each step, the result files are located at the corresponding location.
For example, we start a calculation at `/home/user1/metal_Sn/1_free_energy/400K-0bar-bcc-t`
For NPT MD simulation, the result file will locate at `/home/user1/metal_Sn/1_free_energy/400K-0bar-bcc-t/NPT_sim/result`
For TI simulation the result will locate at `/home/fengbo/4_Sn/1_free_energy/400K-0bar-bct-t/TI_sim/result`
You may want to use the result file and datas of TI_sim/result and plot the free_energy vs T curve for different structure and find the crossing point.
For HTI simulation the result will locate at `/home/fengbo/4_Sn/1_free_energy/400K-0bar-bct-t/HTI_sim/result`, a pure txt file.
The hti out file at
`new_job/02.spring_off/hti.out` which records the integration node on the path is also helpful.
# 💎The procedure of the free energy calculations
To calculate out Gibbs (or Helmholtz) free energy of the materials, there are four steps.
1. NPT MD simulation
1. NVT MD simulation
1. Hamiltonian thermodynamic integration
4. thermodynamic integration
## NPT simulation
Run a long MD simulation and we will get the lattice constant and the best simulation box for the simulations next.
## NVT simulation
Run a long MD simulation with the end structure of NPT simulations. We will know whether the box is reasonable enough from the results of this MD simulation.
## Hamiltonian thermodynamic integration (HTI)
We will know the Gibbs (or Helmholtz) free energy at the specific temperature or pressure condition.
## thermodynamic integration (TI)
Integrating along the isothermal or isobaric path, We will know the free energy at different pressure and temperature.
There are diffefrent json files desinged for different usage.
1. FreeEnergy.json control the whole workflow
1. npt.json
3. nvt.json
3. hti.json or hti.liquid.json
3. ti.t.json or ti.p.json
## FreeEnergy.json
FreeEnergy calculation settings for solid
| Field | Type | Example | Discription |
| --- | --- | --- | --- |
| target_temp | positive integer | 200 | the temperature of HTI |
| target_press | non-negative integer | 50000 | unit :bar.the pressure of HTI |
| work_base_dir | string | "/home/user1/metal_Sn" | see note1. work directory. |
| ti_path | "t" or "p" | "t" | thermodynamic integration along temperature or pressure |
| conf_lmp | string | "bct.lmp" | see note1. the materials structure to be calculated |
| ens | string | "npt-iso" | MD simulation ensemble in lammps |
| if_liquid | bool | false | if simulate liquid |
note:
1. the conf_lmp file must be in the work_base_dir.
the settings used in MD NPT simulations.
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| equi_conf | string | "conf.lmp" | do not change this pair |
| model | string | "graph.pb" | do not change this pair |
| mass_map | list of float | [118.71] | relative atom mass |
| nstep | integer | 1000000 | MD simulation steps in the lammps NPT simulation |
| timestep | float | 0.002 | lammps script time_step. unit: picosecond |
| ens | string | "npt-iso" | lammps MD ensemble setting |
| pres | positive integer | 50000 | pressure in MD simulation (unit:bar; 1GPa==10000bar) |
| tau_t | float | 0.2 | Tdamp in lammps fix npt command |
| tau_p | float | 2.0 | Pdamp in lammps fix npt command |
| thermo_freq | positive integer | 10 | statistic frequency |
## nvt.json
the settings used in MD NPT simulations
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| equi_conf | string | "conf.lmp" | do not change this pair |
| model | string | "graph.pb" | do not change this pair |
| mass_map | list of float | [118.71] | relative atom mass |
| nstep | integer | 1000000 | MD simulation steps in the lammps NPT simulation |
| timestep | float | 0.002 | lammps script time_step. unit: picosecond |
| ens | string | "npt-iso" | lammps MD ensemble setting |
| pres | positive integer | 50000 | pressure in MD simulation (unit:bar; 1GPa==10000bar) |
| tau_t | float | 0.2 | Tdamp in lammps fix npt command |
| tau_p | float | 2.0 | Pdamp in lammps fix npt command |
| thermo_freq | positive integer | 10 | statistic frequency |
## hti.json
For solid, the settings used in Hamiltonian thermodyniamics integration (HTI)
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| equi_conf | string | "conf.lmp" | do not change this pair |
| ncopies | list of integer | [1,1,1] | do not change this pair |
| lambda_lj_on | list of arange | ["0.000:0.100:0.0125",
"0.100:0.200:0.025",
"0.200:1.000:0.2",
"1"] | the lambda value used in 00.lj_on numerial integration |
| lambda_deep_on | list of arange | ["0.00:0.05:0.010",
"0.05:0.15:0.02",
"0.15:0.35:0.040",
"0.35:1.00:0.065",
"1"] | the lambda value used in 01.deep_on numerial integration |
| lambda_spring_off | list of arange | ["0.000:0.750:0.125",
"0.750:0.900:0.050",
"0.900:0.960:0.020",
"0.960:1.00:0.010",
"1"] | the lambda value used in 02.spring_off numerial integration |
| protect_eps | float (usuall small positive number) | 1e-06 | the minimum lambda number used in numerial integration |
| model | string | "graph.pb" | do not change this pair |
| mass_map | list of float | [118.71] | relative atomic mass |
| spring_k | float | 0.02 | spring constant used in Einstein solid. |
| soft_param | dictionary | {"sigma_0_0":2,7,
"epsilon":0.030,
"activation":0.5,
"n":1.0,
"alpha_lj":0.5,
"rcut":6.0} | see: note1 below |
| crystal | "frenkel" or "vega | "frenkel" | different Einstein solid approximation method |
| langevin | bool | true | whether use langevin thermostat |
| nsteps | integer | 200000 | MD steps in each simulation |
| timestep | float | 0.002 | time_step in lammps MD simulation (unit: picosecond) |
| thermo_freq | integer | 10 | statistic frequency(lammps keywork `thermo`) |
| stat_skip | integer | 10000 | skip the first n steps in statistic |
| stat_bsize | integer | 200 | batch size in statistic |
| temp | integer | 400 | the target temperature in HTI calculation |
note:
1. the parameter defined by lammps pair_style lj/cut/soft and pair_coeff command. see [lammps lj/cut/soft](https://lammps.sandia.gov/doc/pair_fep_soft.html#)
2. sigma_0_0 means the sigma value for the lammps atom type 0 and atom type 0.
## hti.liquid.json
For solid, the settings used in Hamiltonian thermodyniamics integration (HTI).
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| equi_conf | string | "conf.lmp" | do not change this pair |
| ncopies | list of integer | [1,1,1] | do not change this pair |
| lambda_soft_on | list of arange | ["0.000:0.030:0.003",
"0.030:0.060:0.005",
"0.060:0.160:0.010",
"0.160:0.300:0.020",
"0.300:1.000:0.050",
"1"] | the lambda value used in 00.soft_on numerial integration |
| lambda_deep_on | list of arange | ["0.000:0.006:0.002",
"0.006:0.030:0.004",
"0.030:0.100:0.010",
"0.100:0.400:0.030",
"0.400:1.000:0.060",
"1"] | the lambda value used in 01.deep_on numerial integration |
| lambda_soft_off | list of arange | ["0.000:0.750:0.125",
"0.750:0.900:0.050",
"0.900:1.000:0.020",
"1"] | the lambda value used in 02.soft_off numerial integration |
| protect_eps | float (usually small positive number) | 1e-06 | the minimum lambda number used in numerial integration |
| model | string | "graph.pb" | do not change this pair |
| mass_map | list of float | [118.71] | relative atomic mass |
| spring_k | float | 0.02 | spring constant used in Einstein solid. |
| soft_param | dictionary | {"sigma_0_0":2,7,
"epsilon":0.030,
"activation":0.5,
"n":1.0,
"alpha_lj":0.5,
"rcut":6.0} | see: note1 below |
| crystal | "frenkel" or "vega | "frenkel" | different Einstein solid approximation method |
| langevin | bool | true | whether use langevin thermostat |
| nsteps | integer | 200000 | MD steps in each simulation |
| timestep | float | 0.002 | time_step in lammps MD simulation (unit: picosecond) |
| thermo_freq | integer | 10 | statistic frequency |
| stat_skip | integer | 10000 | skip the first n steps in statistic |
| stat_bsize | integer | 200 | batch size in statistic |
| temp | integer | 400 | the target temperature in HTI calculation |
note:
1. the parameter defined by lammps pair_style lj/cut/soft and pair_coeff command. see [lammps lj/cut/soft](https://lammps.sandia.gov/doc/pair_fep_soft.html#)
2. sigma_0_0 means the sigma value for the lammps atom type 0 and atom type 0.
## ti.t.json
the settings used in thermodynamic integration (TI) for constant pressure and changeable temperature
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| equi_conf | string | "conf.lmp" | do not change this pair |
| copies | list of integer | [1,1,1] | do not change this pair |
| model | string | "graph.pb" | do not change this pair |
| mass_map | list of float | [118.71] | relative atom mass |
| nstep | integer | 200000 | MD simulation steps in the lammps NPT simulation |
| timestep | float | 0.002 | lammps script time_step. unit: picosecond |
| ens | string | npt-aniso | lammps MD simulation ensemble setting |
| path | "t" or "p" | "t" | do not change this pair for ti.t.json |
| temp_seq | list of arange | ["200:1400:20",
1400] | temperature list to be calculated. The HTI tar_temp must be in it. |
| pres | integer | 50000 | the target pressure of HTI calculation |
| tau_t | float | 0.2 | lammps Tdamp |
| tau_p | float | 2.0 | lammps Pdamp |
| thermo_freq | integer | 10 | statistic frequency |
| stat_skip | integer | 5000 | skip the first n steps in statistic |
| stat_bsize | integer | 200 | statistic batch size |
#
## ti.p.json
the settings used in thermodynamic integration (TI) for constant temperature and changeable pressure
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| equi_conf | string | "conf.lmp" | do not change this pair |
| copies | list of integer | [1,1,1] | do not change this pair |
| model | string | "graph.pb" | do not change this pair |
| mass_map | list of float | [118.71] | relative atom mass |
| nstep | integer | 200000 | MD simulation steps in the lammps NPT simulation |
| timestep | float | 0.002 | lammps script time_step. unit: picosecond |
| ens | string | npt-aniso | lammps MD simulation ensemble setting |
| path | "t" or "p" | "t" | do not change this pair for ti.t.json |
| temp | integer | 800 | temperature to be calculated |
| pres_seq | list of arange | [0:100000:2000,
100000] | the pressure list to be calculated. The HTI tar_pres must be in it. |
| tau_t | float | 0.2 | lammps Tdamp |
| tau_p | float | 2.0 | lammps Pdamp |
| thermo_freq | integer | 10 | statistic frequency |
| stat_skip | integer | 5000 | skip the first n steps in statistic |
| stat_bsize | integer | 200 | statistic batch size |
## gdi.json
The gdi.json is used for gibbs-duham integration. When you know the one point at the two phase coexisting-line. You can do gibbs-duham integration to get the whole phase boundry
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| phase_i | dict | {"name": "PHASE_0",
"equi_conf":"bct.lmp",
"ens":"npt-xy"} | phase 1 information
|
| phase_ii | dict | {"name": "PHASE_1",
"equi_conf":"liquid.lmp",
"ens":"npt-iso"} | phase 2 information |
| model | str | "graph.pb" | |
| mass_map | list of float | [118.71] | relative atomic mass |
| nsteps | integer | 100000 | MD steps in simulation |
| timestep | float | 0.002 | MD timestep (in ps) |
| tau_t | float | 0.1 | MD NPT tau_t |
| tau_p | float | 1.0 | MD NPT tau_p |
| thermo_freq | integer | 10 | MD thermo frequency |
| stat_skip | integer | 5000 | skip the first 5000 value in lammps log |
| stat_bsize | integer | 100 | statistic batch size |
## thermo_freq, stat_bsize, stat_skip example:
if we set nsteps==500000, thermo_freq==10, stat_bsize==100, stat_skip==1000,
then first we will run a 500000 steps MD, and generate 500000/10==50000 data points.
the first stat_skip==1000 points will be ignored,
then the last 49000 points will be grouped into 49000/100==490 chunks.
The average value the of the 100 point value in each chunk will treated as chunk value.
And the 490 chunk values will also be averaged as the final result value for this MD.
### FreeEnergy.json
The json file controls the whole calculation process.
And for each step, it uses corresponding `npt.json` `nvt.json` `hti.json` `ti.json`
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| target_temp | integer | 200 | target simulation temperature unit:K |
| target_pres | integer | 20000 | target simulation pressure unit:Bar.(20000Bar==2GPa) |
| work_base_dir | string | "/home/fengbo/4_Sn/14_free_energy_airflow_test" | should be changed to the dir will this file locates |
| ti_path | string | "t" or "p" | indicate protocol used inthis calculation in TI calculation(along t keep p; along p keep t) |
| conf_lmp | string | "beta.lmp" | structure file name |
| ens | choice:"npt-iso", "npt-aniso", "npt-xy" | 0.002 | the ensemble used in NPT and TI simulation. (solid:npt-aniso; liquid:npt-iso) |
| if_liquid | string | bool | true for liquid, false for other |
### pb.json
example command:
```python gdi.py pb.josn machine.json -g gdidata.json```
This file is used by `dpti gdi` module
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| equi_conf | string | "beta.lmp" | target simulation temperature unit:K |
| ens | choice: "npt-aniso" "npt-iso" "npt-xy" | "npt-xy" | simulation ensemble |
| model | string | "../graph.pb" | model path |
### gdidata.json
This file is used to specify the values GDI (phase coexisting line simulation)
This example shows begining phase transition point at 1GPa,270K. And we want to extend to 1.35GPa .
>Note: when calculating, the solver keeps the local error estimates less than `atol + rtol * abs(y)`. See:[scipy.integrate.solve_ivp doc](https://docs.scipy.org/doc/scipy/reference/generated/scipy.integrate.solve_ivp.html)
Note: if `direction` defines GDI:calculation behaviour.
"p": changing pressure from `begin` to `end` with initial temperature `initial_value`.
"t": changing temperature from `begin` to `end` with initial pressure `initial_value`.
| Field | Type | Example | Description |
| --- | --- | --- | --- |
| begin | integer | 10000 | begin simulation temperature or pressure. unit:K/Bar |
| end | integer | 13500 | end simulation temperature or pressure. unit:K/Bar |
| initial_value | integer | 270 | given initial value |
| abs_tol | float | 2 | used by scipy solve_ivp function |
| rel_tol | float | 0.001 | used by scipy solve_ivp function. |
| direction | choice:"t" "p"| "p" | see above |
# FAQ
## simulation related:
### phase transition:
For thermodynamics integration: there is a restrict that no phase transition should happen during the integration path.
This means that for the simulation process lambda changing from 0 to 1, the structure of system should not change dramatically. (like metal to liquid, lattice constants largely change).
In practice, we could monitor the RDF(compute rdf) of the structure during simulation. The rdf could be used as an indicator for the structure changes. The RMSD value of this system could also be used as indicator.
### integration path:
For thermodynamics integration, sometimes it may not be a choice to directly change from the initial state(Einstein solid, Ideal Gas) to the final target state.
To extend the integration and avoid phase transition during MD simulation, it may be better to introduce some intermediate during simulation, this is implemented in dpti software called two-steps and three-steps.(At least, this strategy could take effects for water(ice) and metal Tin(Sn))
For researcher, it is recommended to try both the direct path protocol and the intermediate state protocol. And compare the results.
And repeat the calculation at least for one more time at a specific temperature and pressure and check the result consistence.
If the result errors lie in about 1meV/per atom. (maybe about 10K-20K in phase diagram) We could treat it as a reliable result.
### atom number in simulation:
Usually the atom number should be about 100-200. Larger system is OK.
Size effect is not obvious.(increasing the simulation size will usually get similar free energy values).