https://github.com/dipterix/filearray
Out-of-memory Arrays in R
https://github.com/dipterix/filearray
array big-data memory-map out-of-memory outofmemory r
Last synced: 7 months ago
JSON representation
Out-of-memory Arrays in R
- Host: GitHub
- URL: https://github.com/dipterix/filearray
- Owner: dipterix
- Created: 2021-09-05T15:58:50.000Z (over 4 years ago)
- Default Branch: main
- Last Pushed: 2024-11-08T16:36:39.000Z (over 1 year ago)
- Last Synced: 2024-12-12T01:47:00.813Z (about 1 year ago)
- Topics: array, big-data, memory-map, out-of-memory, outofmemory, r
- Language: C++
- Homepage: https://dipterix.org/filearray/
- Size: 3.08 MB
- Stars: 17
- Watchers: 1
- Forks: 2
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- Changelog: NEWS.md
Awesome Lists containing this project
README

# File-Backed Array for Out-of-memory Computation
[](https://github.com/dipterix/filearray/actions/workflows/R-CMD-check.yaml)
[](https://CRAN.R-project.org/package=filearray)
[](https://dipterix.r-universe.dev/)
Stores large arrays in files to avoid occupying large memories. Implemented with super fast gigabyte-level multi-threaded reading/writing via `OpenMP`. Supports multiple non-character data types (double, float, integer, complex, logical and raw).
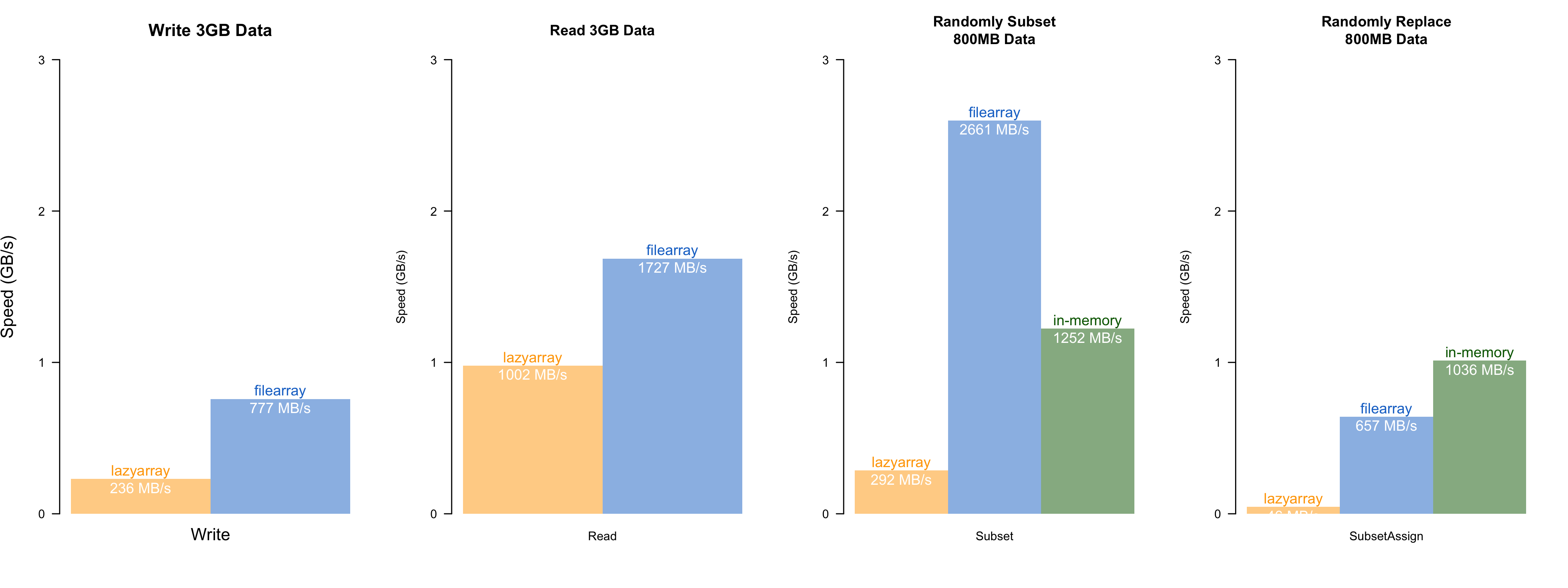
Speed comparisons with `lazyarray` (`zstd`-compressed out-of-memory array), and in-memory operation. The speed test was conducted on an `MacBook Air (M1, 2020, 8GB RAM)`, with 8-threads. `filearray` is uniformly faster than `lazyarray`. Random access has almost the same speed as the native array operation in R. *(The actual speed may vary depending on the storage type and memory size)*
## Installation
```r
install.packages("filearray")
```
### Install Develop Version
The internal functions are written in `C++`. To avoid compiling the packages, you can install from my personal repository. It's automatically updated every hour. Currently available on `Windows` and `osx (Intel chip)` only.
```r
options(repos = c(
dipterix = 'https://dipterix.r-universe.dev',
CRAN = 'https://cloud.r-project.org'))
install.packages('filearray')
```
Alternatively, you can compile from `Github` repository. This requires proper compilers (`rtools` on `windows`, or `xcode-select --install` on `osx`, or `build-essentials` on `linux`).
```r
# install.packages("remotes")
remotes::install_github("dipterix/filearray")
```
## Basic Usage
#### Create/load file array
```r
library(filearray)
file <- tempfile()
x <- filearray_create(file, c(100, 100, 100, 100))
# load existing
x <- filearray_load(file)
```
See more: `help("filearray")`
#### Assign & subset array
```r
x[,,,1] <- rnorm(1e6)
x[1:10,1,1,1]
```
#### Generics
```r
typeof(x)
max(x, na.rm = TRUE)
apply(x, 3, min, na.rm = TRUE)
val = x[1,1,5,1]
fwhich(x, val, arr.ind = TRUE)
```
See more: `help("S3-filearray")`, `help("fwhich")`
#### Map-reduce
Process segments of array and reduce to save memories.
```
# Identical to sum(x, na.rm = TRUE)
mapreduce(x,
map = \(data){ sum(data, na.rm = TRUE) },
reduce = \(mapped){ do.call(sum, mapped) })
```
See more: `help("mapreduce")`
#### Collapse
Transform data, and collapse (calculate sum or mean) along margins.
```
a <- x$collapse(keep = 4, method = "mean", transform = "asis")
# equivalent to
b <- apply(x[], 4, mean)
a[1] - b[1]
```
Available `transform` for double/integer numbers are:
* `asis`: no transform
* `10log10`: `10 * log10(v)`
* `square`: `v * v`
* `sqrt`: `sqrt(v)`
For complex numbers, `transform` is a little bit different:
* `asis`: no transform
* `10log10`: `10 * log10(|x|^2)` (power to decibel unit)
* `square`: `|x|^2`
* `sqrt`: `|x|` (modulus)
* `normalize`: `x / |x|` (unit length)
## Notes
#### I. Notes on precision
1. `complex` numbers: In native `R`, complex numbers are combination of two `double` numbers - real and imaginary (total 16 bytes). In `filearray`, complex numbers are coerced to two `float` numbers and store each number in 8 bytes. This conversion will gain performance speed, but lose precision at around 8 decimal place. For example, `1.0000001` will be store as `1`, or `123456789` will be stored as `123456792` (first 7 digits are accurate).
2. `float` type: Native R does not have float type. All numeric values are stored in double precision. Since float numbers use half of the space, float arrays can be faster when hard drive speed is the bottle-neck (see [performance comparisons](https://dipterix.org/filearray/articles/performance.html)). However coercing double to float comes at costs:
a). float number has less precision
b). float number has smaller range ($3.4\times 10^{38}$) than double ($1.7\times 10^{308}$)
hence use with caution when data needs high precision or the max is super large.
3. `collapse` function: when data range is large (say `x[[1]]=1`, but `x[[2]]=10^20`), `collapse` method might lose precision. This is `double` only uses 8 bytes of memory space. When calculating summations, R internally uses `long double` to prevent precision loss, but current `filearray` implementation uses `double`, causing floating error around 16 decimal place.
#### II. Cold-start vs warm-start
As of version `0.1.1`, most file read/write operations are switched from `fopen` to memory map for two simplify the logic (buffer size, kernel cache...), and to boost the writing/some types of reading speed. While sacrificing the speed of reading large block of data from 2.4GB/s to 1.7GB/s, the writing speed was boosted from 300MB/s to 700MB/s, and the speed of random accessing small slices of data was increased from 900MB/s to 2.5GB/s. As a result, some functions can reach to really high speed (close to in-memory calls) while using much less memory.
The additional performance improvements brought by the memory mapping approach might be impacted by "cold" start. When reading/writing files, most modern systems will cache the files so that it can load up these files faster next time. I personally call it a cold start. Memory mapping have a little bit extra overhead during the cold start, resulting in decreased performance (but it's still fast). Accessing the same data after the cold start is called warm start. When operating with warm starts, `filearray` is as fast as native R arrays (sometimes even faster due to the indexing method and fewer garbage collections). This means `filearray` reaches its best performance when the arrays are re-used.
#### III. Using traditional HDD?
`filearray` relies on `SSD`, especially `NVMe SSD` that allows you to fast-access random hard disk address. If you use `HDD`, `filearray` can provide very limited improvement.
If you use `filearray` to direct access to HDD, please set number of threads to `1` via `filearray::filearray_threads(1)` at start up, or set system environment `FILEARRAY_NUM_THREADS` to `"1"`.