Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/dkratzert/dsr
Refinement of disordered structures with SHELXL
https://github.com/dkratzert/dsr
crystallography database dsr fragments molecule python shelxl
Last synced: 3 months ago
JSON representation
Refinement of disordered structures with SHELXL
- Host: GitHub
- URL: https://github.com/dkratzert/dsr
- Owner: dkratzert
- Created: 2014-02-01T19:38:00.000Z (about 11 years ago)
- Default Branch: master
- Last Pushed: 2024-06-01T09:04:28.000Z (8 months ago)
- Last Synced: 2024-10-11T18:07:49.534Z (4 months ago)
- Topics: crystallography, database, dsr, fragments, molecule, python, shelxl
- Language: Python
- Homepage: https://dkratzert.de/dsr.html
- Size: 475 MB
- Stars: 5
- Watchers: 2
- Forks: 1
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- Funding: .github/FUNDING.yml
Awesome Lists containing this project
README

[](https://github.com/dkratzert/DSR/actions/workflows/pythonapp.yml)

[](https://repology.org/project/python:dsr-shelx/versions)
DSR
===
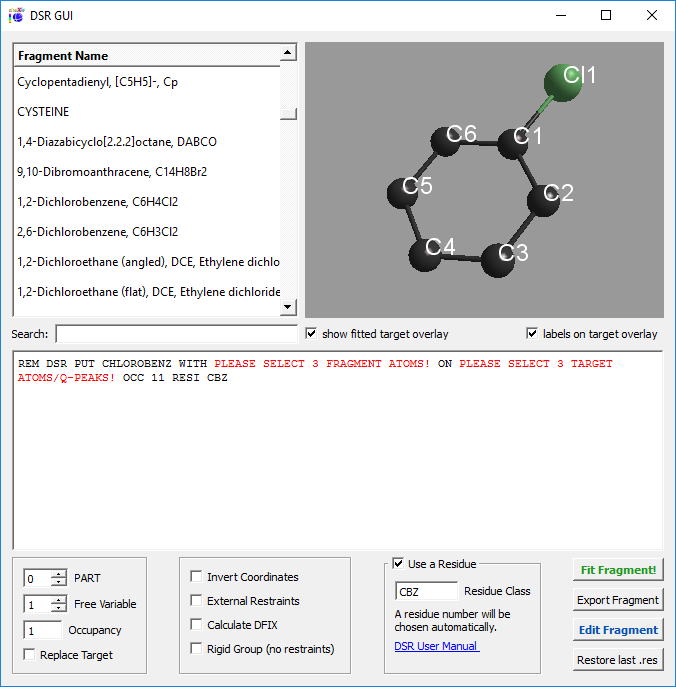
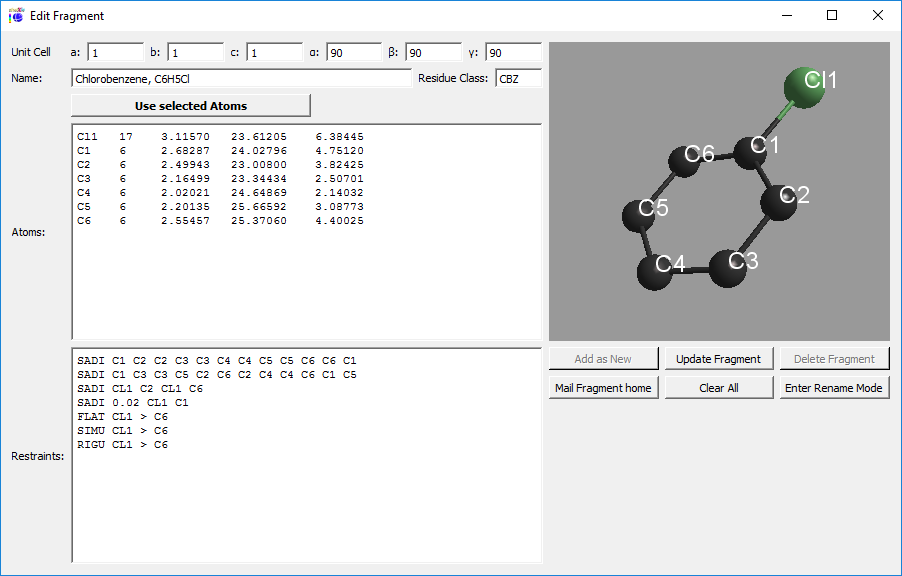
The program DSR consists of a text database with fragments of molecules and the DSR program.
It acts as a preprocessor for SHELXL .res files. The user inserts a special command in the SHELXL .res file
and the DSR program reads this information to put a molecule or fragment with the desired atoms on the position
of the target atoms or q-peaks in the unit cell. Bond restraints can be either applied from the database to the molecule
or automatically generated.
* [The homepage](https://dkratzert.de/dsr.html)
I apologize for the messy code. This was my first bigger project...
You have either a command line version:
```
C:\Users\daniel>dsr
----------------------------------------------------- D S R - v238 -------------------------------------
Disordered Structure Refinement (DSR)
Example DSR .res file command line:
REM DSR PUT/REPLACE "Fragment" WITH C1 C2 C3 ON Q1 Q2 Q3 PART 1 OCC -21 =
RESI DFIX
---------------------------------------------------------------------------------------------------------
PUT: Just put the fragment source atoms here.
REPLACE: Replace atoms of PART 0 in 1.3 A distance around target atoms.
---------------------------------------------------------------------------------------------------------
optional arguments:
-h, --help show this help message and exit
-r "res file" ["res file" ...]
res file with DSR command
-re "res file" ["res file" ...]
res file with DSR command (write restraints to external file)
-e "fragment" export fragment as .res/.png file
-c "fragment" export fragment to clipboard
-t inverts the current fragment
-i "tgz file" import a fragment from GRADE (needs .tgz file)
-l list names of all database entries
-s "string" search the database for a name
-g keep group rigid (no restraints)
-u Update DSR to the most current version
-n do not refine after fragment transfer
```
Or a graphical user interface in [ShelXle](https://www.shelxle.org/shelx/eingabe.php):