https://github.com/dnth/microalgae-cell-counter-blogpost
Accompanying repo for the blogpost Modelling a Microalgae Cell Counter with Deep Learning on dicksonneoh.com
https://github.com/dnth/microalgae-cell-counter-blogpost
Last synced: about 2 months ago
JSON representation
Accompanying repo for the blogpost Modelling a Microalgae Cell Counter with Deep Learning on dicksonneoh.com
- Host: GitHub
- URL: https://github.com/dnth/microalgae-cell-counter-blogpost
- Owner: dnth
- Created: 2022-04-07T06:44:02.000Z (about 3 years ago)
- Default Branch: main
- Last Pushed: 2022-06-15T05:24:29.000Z (almost 3 years ago)
- Last Synced: 2025-03-25T18:12:22.424Z (about 2 months ago)
- Language: Jupyter Notebook
- Size: 116 MB
- Stars: 1
- Watchers: 1
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
Awesome Lists containing this project
README
# Training a Deep Learning Model for Cell Counting in 17 Lines of Code with 17 Images
This is an accompanying repo for the blog post Training a Deep Learning Model for Cell Counting in 17 Lines of Code with 17 Images.
Link to the full blog post
https://dicksonneoh.com/portfolio/training_dl_model_for_cell_counting/
In this blog post, I will walk you through how to use the IceVision library and train a state-of-the-art deep learning model with Fastai to count microalgae cells.
Among the things you will learn:
* Installation of the libraries.
* Prepare and label any dataset for object detection.
* Train a high-performance VFNet model with IceVision & Fastai.
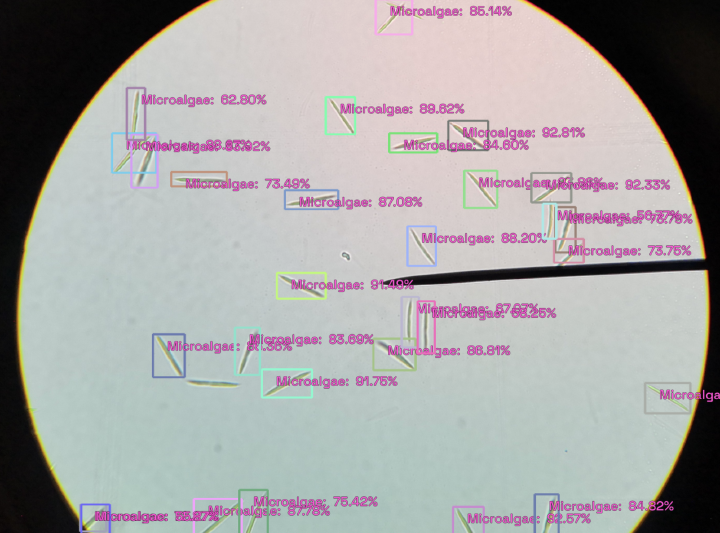
* Use the model for inference on new images.
By the end of the post, you will have an object detection model that will automatically detect microalgae cells from an image.
# Support Me
If you like what you see, support me in keeping the lights on to produce more posts like this.
