https://github.com/dylanljones/lattpy
Simple and efficient Python package for modeling d-dimensional Bravais lattices in solid state physics.
https://github.com/dylanljones/lattpy
bravais bravais-lattice computational-physics crystal high-dimensional lattice nearest-neighbors neighbors physics python solid-state-physics
Last synced: 10 months ago
JSON representation
Simple and efficient Python package for modeling d-dimensional Bravais lattices in solid state physics.
- Host: GitHub
- URL: https://github.com/dylanljones/lattpy
- Owner: dylanljones
- License: mit
- Created: 2020-04-22T08:46:36.000Z (almost 6 years ago)
- Default Branch: master
- Last Pushed: 2025-02-17T19:42:26.000Z (12 months ago)
- Last Synced: 2025-03-31T04:08:10.085Z (11 months ago)
- Topics: bravais, bravais-lattice, computational-physics, crystal, high-dimensional, lattice, nearest-neighbors, neighbors, physics, python, solid-state-physics
- Language: Python
- Homepage: https://lattpy.readthedocs.io/en/stable/
- Size: 1.45 MB
- Stars: 39
- Watchers: 1
- Forks: 12
- Open Issues: 3
-
Metadata Files:
- Readme: README.md
- Changelog: CHANGELOG.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
- Citation: CITATION.cff
Awesome Lists containing this project
README
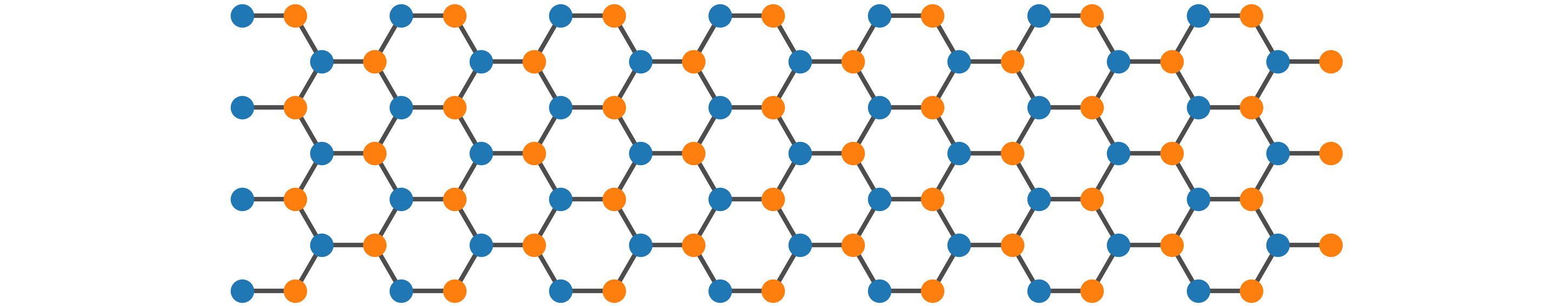
LattPy - Simple Lattice Modeling in Python
:warning: **WARNING**: This project is still in development and might change significantly in the future!
*LattPy* is a simple and efficient Python package for modeling Bravais lattices and
constructing (finite) lattice structures in any dimension.
It provides an easy interface for constructing lattice structures by simplifying the
configuration of the unit cell and the neighbor connections - making it possible to
construct complex models in just a few lines of code and without the headache of
adding neighbor connections manually. You will save time and mental energy for more important matters.
| Master | [![Test][tests-master]][link-tests] | [![Codecov][codecov-master]][codecov-master-link] | [![Read the Docs][docs-master]][docs-master-link] |
|:-------|:------------------------------------|:--------------------------------------------------|:--------------------------------------------------|
| Dev | [![Test][tests-dev]][link-tests] | [![Codecov][codecov-dev]][codecov-dev-link] | [![Read the Docs][docs-dev]][docs-dev-link] |
## 🔧 Installation
LattPy is available on [PyPI](https://pypi.org/project/lattpy/):
````commandline
pip install lattpy
````
Alternatively, it can be installed via [GitHub](https://github.com/dylanljones/lattpy)
```commandline
pip install git+https://github.com/dylanljones/lattpy.git@VERSION
```
where `VERSION` is a release or tag. The project can also be
cloned/forked and installed via
````commandline
python setup.py install
````
## 📖 Documentation
[Read the documentation on ReadTheDocs!][docs-stable-link]
## 🚀 Quick-Start
See the [tutorial][docs-tutorial-link] for more information and examples.
Features:
- Basis transformations
- Configurable unit cell
- Easy neighbor configuration
- General lattice structures
- Finite lattice models in world or lattice coordinates
- Periodic boundary conditions along any axis
### Configuration
A new instance of a lattice model is initialized using the unit-vectors of the Bravais lattice.
After the initialization the atoms of the unit-cell need to be added. To finish the configuration
the connections between the atoms in the lattice have to be set. This can either be done for
each atom-pair individually by calling ``add_connection`` or for all possible pairs at once by
callling ``add_connections``. The argument is the number of unique
distances of neighbors. Setting a value of ``1`` will compute only the nearest
neighbors of the atom.
````python
import numpy as np
from lattpy import Lattice
latt = Lattice(np.eye(2)) # Construct a Bravais lattice with square unit-vectors
latt.add_atom(pos=[0.0, 0.0]) # Add an Atom to the unit cell of the lattice
latt.add_connections(1) # Set the maximum number of distances between all atoms
latt = Lattice(np.eye(2)) # Construct a Bravais lattice with square unit-vectors
latt.add_atom(pos=[0.0, 0.0], atom="A") # Add an Atom to the unit cell of the lattice
latt.add_atom(pos=[0.5, 0.5], atom="B") # Add an Atom to the unit cell of the lattice
latt.add_connection("A", "A", 1) # Set the max number of distances between A and A
latt.add_connection("A", "B", 1) # Set the max number of distances between A and B
latt.add_connection("B", "B", 1) # Set the max number of distances between B and B
latt.analyze()
````
Configuring all connections using the ``add_connections``-method will call the ``analyze``-method
directly. Otherwise this has to be called at the end of the lattice setup or by using
``analyze=True`` in the last call of ``add_connection``. This will compute the number of neighbors,
their distances and their positions for each atom in the unitcell.
To speed up the configuration prefabs of common lattices are included. The previous lattice
can also be created with
````python
from lattpy import simple_square
latt = simple_square(a=1.0, neighbors=1) # Initializes a square lattice with one atom in the unit-cell
````
So far only the lattice structure has been configured. To actually construct a (finite) model of the lattice
the model has to be built:
````python
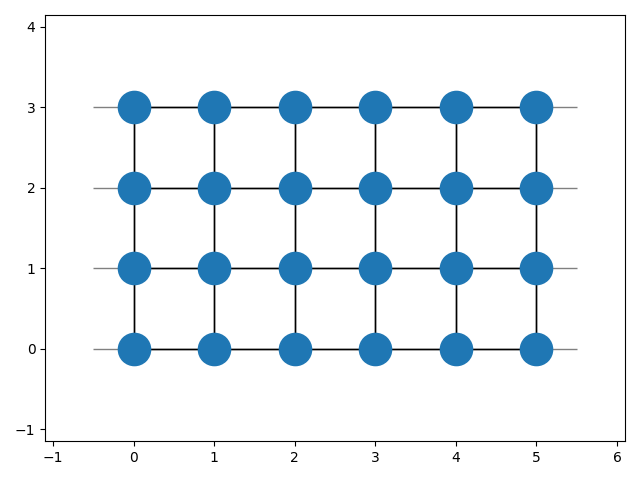
latt.build(shape=(5, 3))
````
This will compute the indices and neighbors of all sites in the given shape and store the data.
After building the lattice periodic boundary conditions can be set along one or multiple axes:
````python
latt.set_periodic(axis=0)
````
To view the built lattice the `plot`-method can be used:
````python
import matplotlib.pyplot as plt
latt.plot()
plt.show()
````
### General lattice attributes
After configuring the lattice the attributes are available.
Even without building a (finite) lattice structure all attributes can be computed on the fly for a given lattice vector,
consisting of the translation vector `n` and the atom index `alpha`. For computing the (translated) atom positions
the `get_position` method is used. Also, the neighbors and the vectors to these neighbors can be calculated.
The `dist_idx`-parameter specifies the distance of the neighbors (0 for nearest neighbors, 1 for next nearest neighbors, ...):
````python
from lattpy import simple_square
latt = simple_square()
# Get position of atom alpha=0 in the translated unit-cell
positions = latt.get_position(n=[0, 0], alpha=0)
# Get lattice-indices of the nearest neighbors of atom alpha=0 in the translated unit-cell
neighbor_indices = latt.get_neighbors(n=[0, 0], alpha=0, distidx=0)
# Get vectors to the nearest neighbors of atom alpha=0 in the translated unit-cell
neighbor_vectors = latt.get_neighbor_vectors(alpha=0, distidx=0)
````
Also, the reciprocal lattice vectors can be computed
````python
rvecs = latt.reciprocal_vectors()
````
or used to construct the reciprocal lattice:
````python
rlatt = latt.reciprocal_lattice()
````
The 1. Brillouin zone is the Wigner-Seitz cell of the reciprocal lattice:
````python
bz = rlatt.wigner_seitz_cell()
````
The 1.BZ can also be obtained by calling the explicit method of the direct lattice:
````python
bz = latt.brillouin_zone()
````
### Finite lattice data
If the lattice has been built the needed data is cached. The lattice sites of the
structure then can be accessed by a simple index `i`. The syntax is the same as before,
just without the `get_` prefix:
````python
latt.build((5, 2))
i = 2
# Get position of the atom with index i=2
positions = latt.position(i)
# Get the atom indices of the nearest neighbors of the atom with index i=2
neighbor_indices = latt.neighbors(i, distidx=0)
# the nearest neighbors can also be found by calling (equivalent to dist_idx=0)
neighbor_indices = latt.nearest_neighbors(i)
````
### Data map
The lattice model makes it is easy to construct the (tight-binding) Hamiltonian of a non-interacting model:
````python
import numpy as np
from lattpy import simple_chain
# Initializes a 1D lattice chain with a length of 5 atoms.
latt = simple_chain(a=1.0)
latt.build(shape=4)
n = latt.num_sites
# Construct the non-interacting (kinetic) Hamiltonian-matrix
eps, t = 0., 1.
ham = np.zeros((n, n))
for i in range(n):
ham[i, i] = eps
for j in latt.nearest_neighbors(i):
ham[i, j] = t
````
Since we loop over all sites of the lattice the construction of the hamiltonian is slow.
An alternative way of mapping the lattice data to the hamiltonian is using the `DataMap`
object returned by the `map()` method of the lattice data. This stores the atom-types,
neighbor-pairs and corresponding distances of the lattice sites. Using the built-in
masks the construction of the hamiltonian-data can be vectorized:
````python
from scipy import sparse
# Vectorized construction of the hamiltonian
eps, t = 0., 1.
dmap = latt.data.map() # Build datamap
values = np.zeros(dmap.size) # Initialize array for data of H
values[dmap.onsite(alpha=0)] = eps # Map onsite-energies to array
values[dmap.hopping(distidx=0)] = t # Map hopping-energies to array
# The indices and data array can be used to construct a sparse matrix
ham_s = sparse.csr_matrix((values, dmap.indices))
ham = ham_s.toarray()
````
Both construction methods will create the following Hamiltonian-matrix:
````
[[0. 1. 0. 0. 0.]
[1. 0. 1. 0. 0.]
[0. 1. 0. 1. 0.]
[0. 0. 1. 0. 1.]
[0. 0. 0. 1. 0.]]
````
## 🔥 Performance
Even though `lattpy` is written in pure python, it achieves high performance and
a low memory footprint by making heavy use of numpy's vectorized operations and scipy's
cKDTree. As an example the build times and memory usage in the build process for different
lattices are shown in the following plots:
| Build time | Build memory |
|:-------------------------------------------------------------------------------------------------------:|:---------------------------------------------------------------------------------------------------------:|
|  |
|  |
|
Note that the overhead of the multi-thread neighbor search results in a slight
increase of the build time for small systems. By using `num_jobs=1` in the `build`-method
this overhead can be eliminated for small systems. By passing `num_jobs=-1` all cores
of the system are used.
## 💻 Development
See the [CHANGELOG](https://github.com/dylanljones/lattpy/blob/master/CHANGELOG.md) for
the recent changes of the project.
If you encounter an issue or want to contribute to pyrekordbox, please feel free to
get in touch, [create an issue](https://github.com/dylanljones/lattpy/issues/new/choose)
or open a pull request! A guide for contributing to `lattpy` and the commit-message style can be found in
[CONTRIBUTING](https://github.com/dylanljones/lattpy/blob/master/CONTRIBUTING.md)
[docs-stable-link]: https://lattpy.readthedocs.io/en/stable/
[docs-tutorial-link]: https://lattpy.readthedocs.io/en/stable/tutorial/index.html
[tests-master]: https://img.shields.io/github/actions/workflow/status/dylanljones/lattpy/test.yml?branch=master&label=tests&logo=github&style=flat
[tests-dev]: https://img.shields.io/github/actions/workflow/status/dylanljones/lattpy/test.yml?branch=dev&label=tests&logo=github&style=flat
[link-tests]: https://github.com/dylanljones/lattpy/actions/workflows/test.yml
[codecov-master]: https://codecov.io/gh/dylanljones/lattpy/branch/master/graph/badge.svg?
[codecov-master-link]: https://app.codecov.io/gh/dylanljones/lattpy/branch/master
[codecov-dev]: https://codecov.io/gh/dylanljones/lattpy/branch/dev/graph/badge.svg?
[codecov-dev-link]: https://app.codecov.io/gh/dylanljones/lattpy/branch/dev
[docs-master]: https://img.shields.io/readthedocs/lattpy/latest?style=flat&logo=readthedocs
[docs-master-link]: https://lattpy.readthedocs.io/en/latest/
[docs-dev]: https://img.shields.io/readthedocs/lattpy/dev?style=flat&logo=readthedocs
[docs-dev-link]: https://lattpy.readthedocs.io/en/dev/
[code-ql]: https://github.com/dylanljones/lattpy/actions/workflows/codeql.yml/badge.svg
[code-ql-link]: https://github.com/dylanljones/lattpy/actions/workflows/codeql.yml




