https://github.com/ec-kity/ec-kity
EC-KitY: A scikit-learn-compatible Python tool kit for doing evolutionary computation.
https://github.com/ec-kity/ec-kity
evolutionary-algorithms evolutionary-computation genetic-algorithms genetic-programming machine-learning python scikit-learn sklearn symbolic-regression
Last synced: 11 months ago
JSON representation
EC-KitY: A scikit-learn-compatible Python tool kit for doing evolutionary computation.
- Host: GitHub
- URL: https://github.com/ec-kity/ec-kity
- Owner: EC-KitY
- License: bsd-3-clause
- Created: 2021-11-29T15:09:42.000Z (about 4 years ago)
- Default Branch: develop
- Last Pushed: 2025-01-30T17:29:22.000Z (about 1 year ago)
- Last Synced: 2025-04-04T09:37:37.275Z (11 months ago)
- Topics: evolutionary-algorithms, evolutionary-computation, genetic-algorithms, genetic-programming, machine-learning, python, scikit-learn, sklearn, symbolic-regression
- Language: Python
- Homepage: https://www.eckity.org
- Size: 4.88 MB
- Stars: 93
- Watchers: 4
- Forks: 18
- Open Issues: 25
-
Metadata Files:
- Readme: README.md
- License: LICENSE
- Citation: CITATION.cff
Awesome Lists containing this project
README


**EC-KitY** is a Python tool kit for doing evolutionary computation, and it is scikit-learn compatible.
Currently we have implemented Genetic Algorithm (GA) and tree-based Genetic Programming (GP), but EC-KitY will grow!
[Join the community section](https://chat.eckity.org)
**EC-KitY** is:
- A comprehensive toolkit for running evolutionary algorithms
- Written in Python
- Can work with or without scikit-learn, i.e., supports both sklearn and non-sklearn modes
- Designed with modern software engineering in mind
- Designed to support all popular EC paradigms (GA, GP, ES, coevolution, multi-objective, etc').
### Dependencies
The minimal Python Version for EC-KitY is Python 3.8
The dependencies of our package are described in `requirements.txt`
For sklearn mode, EC-KitY additionally requires:
- scikit-learn (>=1.1)
### User installation
`pip install eckity`
### Documentation
API is available [here](https://api.eckity.org)
(Work in progress - some modules and functions are not documented yet.)
### Tutorials
The tutorials are available [here](https://github.com/EC-KitY/EC-KitY/wiki/Tutorials), walking you through running EC-KitY both in sklearn mode and in non-sklearn mode.
### Examples
More examples are in the [examples](https://github.com/EC-KitY/EC-KitY/tree/main/examples "examples") folder.
All you need to do is define a fitness-evaluation method, through a `SimpleIndividualEvaluator` sub-class.
You can run the examples with ease by opening this [colab notebook](https://colab.research.google.com/drive/1mpr3EGb1rpoK-_zugszQkv1sWVm-ZQiB?usp=sharing).
### Basic example (no sklearn)
You can run an EA with just 3 lines of code. The problem being solved herein is simple symbolic regression.
Additional information on this problem can be found in the [Symbolic Regression Tutorial](https://github.com/EC-KitY/EC-KitY/wiki/Tutorial:-Symbolic-Regression).
```python
from eckity.subpopulation import Subpopulation
from eckity.algorithms import SimpleEvolution
from eckity.base.untyped_functions import f_add, f_sub, f_mul, f_div
from eckity.creators import FullCreator
from eckity.genetic_operators import SubtreeCrossover, SubtreeMutation
from examples.treegp.basic_mode.symbolic_regression import SymbolicRegressionEvaluator
algo = SimpleEvolution(
Subpopulation(
SymbolicRegressionEvaluator(),
creators=FullCreator(
terminal_set=['x', 'y', 'z'],
function_set=[f_add, f_sub, f_mul, f_div]
),
operators_sequence=[SubtreeCrossover(), SubtreeMutation()]
)
)
algo.evolve()
print(f'algo.execute(x=2,y=3,z=4): {algo.execute(x=2, y=3, z=4)}')
```
### Example with sklearn
The problem being solved herein is the same problem, but in this case we also involve sklearn compatability - a core feature of EC-KitY.
Additional information for this example can be found in the [Sklearn Symbolic Regression Tutorial](https://github.com/EC-KitY/EC-KitY/wiki/Tutorial:-Sklearn-Compatible-Symbolic-Regression).
A simple sklearn-compatible EA run:
```python
from sklearn.datasets import make_regression
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from eckity.algorithms.simple_evolution import SimpleEvolution
from eckity.base.untyped_functions import f_add, f_div, f_mul, f_sub
from eckity.creators.gp_creators.full import FullCreator
from eckity.genetic_encodings.gp.tree.utils import create_terminal_set
from eckity.genetic_operators import SubtreeCrossover, SubtreeMutation
from eckity.sklearn_compatible.regression_evaluator import RegressionEvaluator
from eckity.sklearn_compatible.sk_regressor import SKRegressor
from eckity.subpopulation import Subpopulation
X, y = make_regression(n_samples=100, n_features=3)
terminal_set = create_terminal_set(X)
function_set = [f_add, f_sub, f_mul, f_div]
algo = SimpleEvolution(
Subpopulation(
RegressionEvaluator(),
creators=FullCreator(
terminal_set=terminal_set,
function_set=function_set
),
operators_sequence=[SubtreeCrossover(), SubtreeMutation()]
)
)
regressor = SKRegressor(algo)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
regressor.fit(X_train, y_train)
print('MAE on test set:', mean_absolute_error(y_test, regressor.predict(X_test)))
```
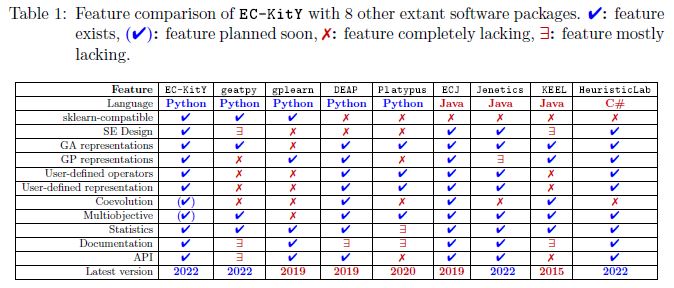
### Feature comparison
Here's a comparison table. The full paper is available [here](https://arxiv.org/abs/2207.10367).
### Authors
[Moshe Sipper](http://www.moshesipper.com/ "Moshe Sipper"),
[Achiya Elyasaf](https://achiya.elyasaf.net/ "Achiya Elyasaf"),
[Itai Tzruia](https://www.linkedin.com/in/itai-tzruia-4a47a91b8/),
Tomer Halperin
### Citation
Citations are always appreciated 😊:
```
@article{eckity2023,
author = {Moshe Sipper and Tomer Halperin and Itai Tzruia and Achiya Elyasaf},
title = {{EC-KitY}: Evolutionary computation tool kit in {Python} with seamless machine learning integration},
journal = {SoftwareX},
volume = {22},
pages = {101381},
year = {2023},
url = {https://www.sciencedirect.com/science/article/pii/S2352711023000778},
}
@misc{eckity2022git,
author = {Sipper, Moshe and Halperin, Tomer and Tzruia, Itai and Elyasaf, Achiya},
title = {{EC-KitY}: Evolutionary Computation Tool Kit in {Python}},
year = {2022},
publisher = {GitHub},
journal = {GitHub repository},
howpublished = {\url{https://www.eckity.org/} }
}
```
### Sample repos using EC-KitY
- [EC-KitY-Maze-Example](https://github.com/RonMichal/EC-KitY-Maze-Example/tree/maze_example/examples/vectorga/maze)
- [EvolutionTSP](https://github.com/nogazax/EvolutionTSP)
- [Solving The 'Nurse Scheduling Problem' With EC-KitY](https://github.com/harelaf/Nurse-Scheduling-Problem)