https://github.com/edgar-mendonca/shpb-analysis
Python based SHPB Experimental data analysis
https://github.com/edgar-mendonca/shpb-analysis
matplotlib numpy pandas python scipy
Last synced: 2 months ago
JSON representation
Python based SHPB Experimental data analysis
- Host: GitHub
- URL: https://github.com/edgar-mendonca/shpb-analysis
- Owner: Edgar-Mendonca
- License: apache-2.0
- Created: 2024-03-25T05:23:27.000Z (about 1 year ago)
- Default Branch: main
- Last Pushed: 2024-04-05T05:20:45.000Z (about 1 year ago)
- Last Synced: 2025-04-11T16:36:35.274Z (2 months ago)
- Topics: matplotlib, numpy, pandas, python, scipy
- Language: Python
- Homepage:
- Size: 1.48 MB
- Stars: 4
- Watchers: 1
- Forks: 1
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# SHPB Analysis Tool
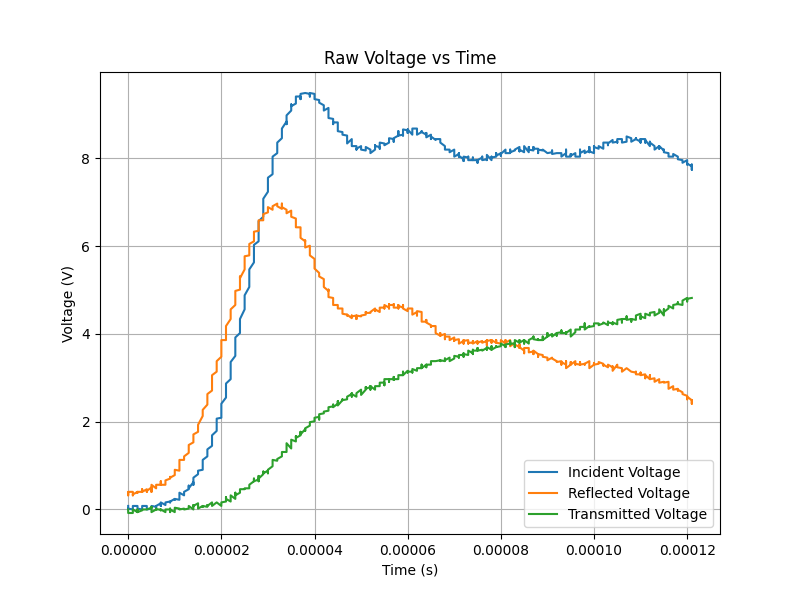
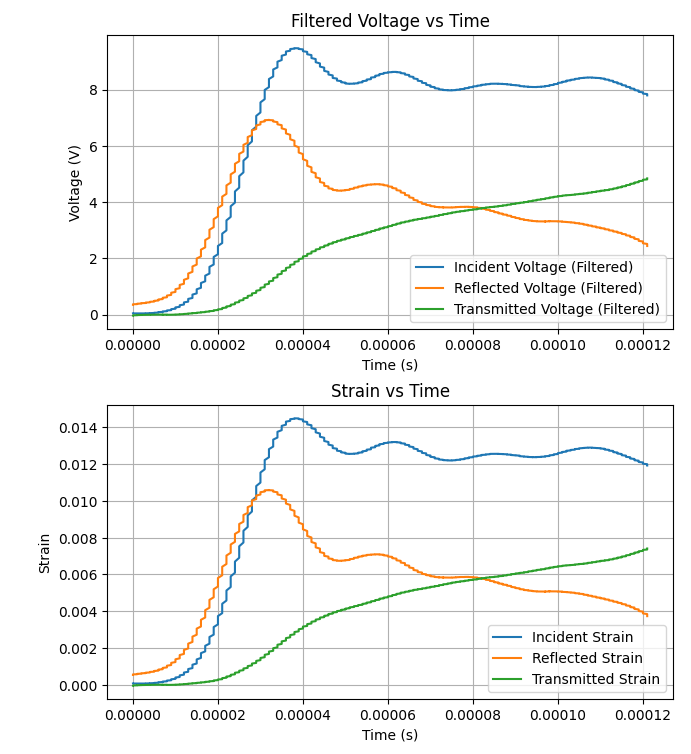
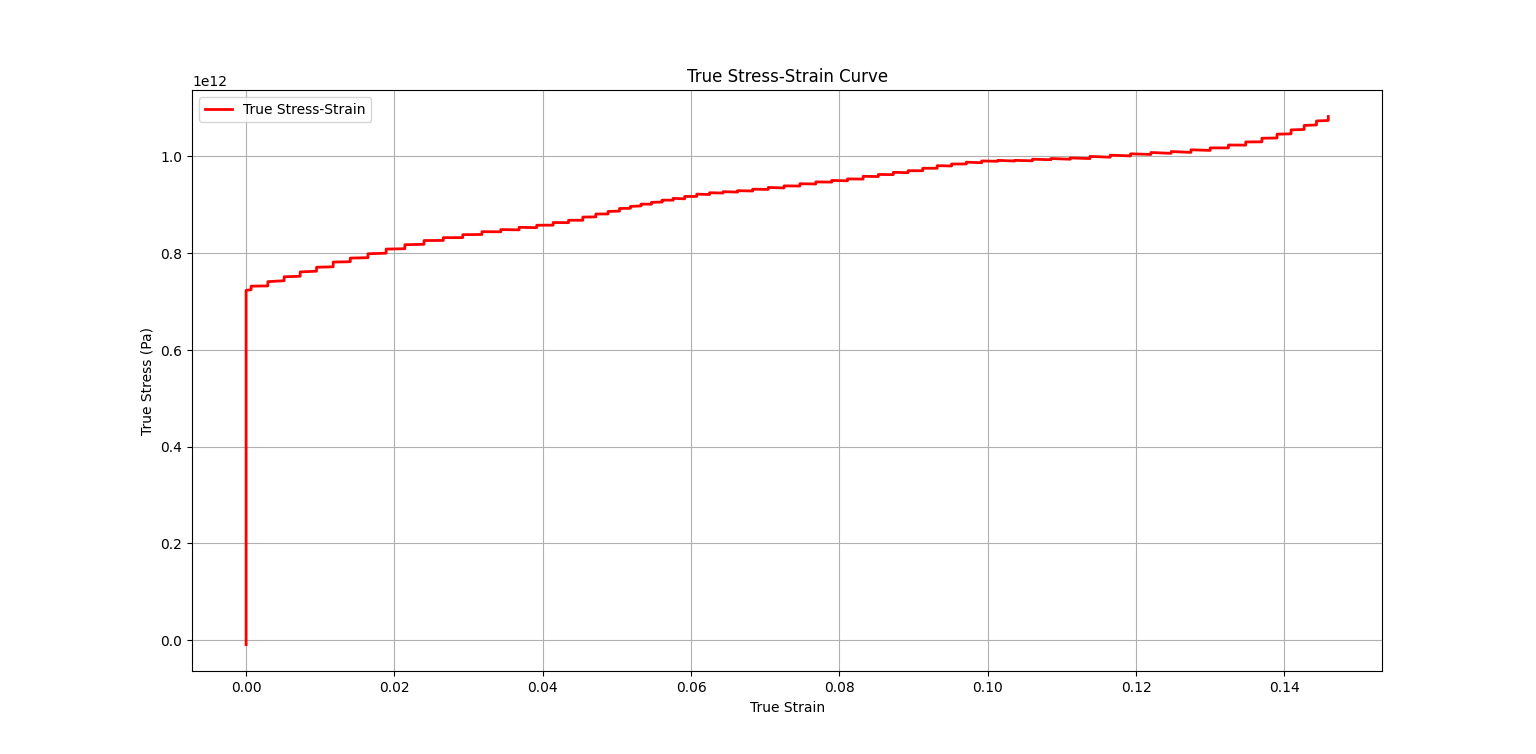
This Python script analyses Split Hopkinson Pressure Bar (SHPB) experimental data to calculate stress-strain curves and other relevant parameters. Follow the instructions below to use the script with your experimental data.
## Instructions
1. **Install Dependencies:** Make sure you have Python installed on your system. You can install the required Python packages by running the following command:
```
pip install -r requirements.txt
```
2. **Prepare Experimental Data:**
- Ensure that your experimental data is stored in a CSV or Excel file.
- The file should contain columns for "Time", "Incident", "Reflected", and "Transmitted" voltages.
- Voltage values should be in volts (V).
- Specify the file name in the Python script (`analysis.py`) using the `filename` variable. For example:
```python
filename = "path/to/your/data_file.csv"
```
3. **Customize Input Parameters:**
- Open the Python script (`analysis.py`) in a text editor.
- Update the input parameters according to your experiment setup. Parameters such as Young's modulus, density, initial length, and cross-sectional areas should be adjusted based on your experimental conditions.
4. **Run the Script:**
- Run the script using the following command:
```
python analysis.py
```
5. **View Plots and Analyze Results:**
- After running the script, view the generated plots to analyze the results.
- The script will generate plots for filtered voltage data, strain data, stress-strain curve, and true stress-strain curve.
## Example Usage
Here's an example of how to use the script with your experimental data:
1. Ensure that Python and the required dependencies are installed on your system.
2. Prepare your experimental data in a CSV or Excel file format.
3. Customize the input parameters in the `analysis.py` script based on your experiment setup.
4. Specify the file name of your experimental data in the script.
5. Run the script using `python analysis.py`.
6. Analyze the generated plots to interpret the results of your SHPB experiment.
## Explanation for Stress-Strain Calculation in SHPB Experiment
In the Split Hopkinson Pressure Bar (SHPB) experiment, the characteristic relations associated with one-dimensional elastic wave propagation in the bar provide the basis for calculating stress and strain in the specimen.
1. **Particle Velocity at Specimen/Input-Bar and Specimen/Output-Bar Interface:**
- The particle velocity $` v_1(t) `$ at the specimen/input-bar interface is given by:
$` v_1(t) = c_b(\varepsilon_I - \varepsilon_R) `$
- Here, $` c_b = \sqrt{\frac{E_b}{\rho_b}} `$ represents the bar wave speed, with $` E_b `$ denoting the Young's modulus and $` \rho_b `$ the density of the bar material.
- The particle velocity $` v_2(t) `$ at the specimen/output-bar interface is given by:
$` v_2(t) = c_b \varepsilon_T `$
2. **Mean Axial Strain Rate in the Specimen:**
- The mean axial strain rate $` \dot{e}_s `$ in the specimen is calculated as:
$` \dot{e}_s = \frac{c_b}{l_0} (\varepsilon_I - \varepsilon_R - \varepsilon_T) = \frac{v_1 - v_2}{l_0} `$
- Here, $` l_0 `$ represents the initial specimen length.
3. **Calculation of Bar Stresses and Normal Forces:**
- The stresses and normal forces at the specimen/bar interfaces are computed as follows:
- $` P_1 = E_b (\varepsilon_I + \varepsilon_R) A_b `$ at the specimen/input-bar interface.
- $` P_2 = E_b \varepsilon_T A_b `$ at the specimen/output-bar interface.
- Here, $` A_b `$ denotes the cross-sectional area of the bars.
4. **Mean Axial Stress in the Specimen:**
- The mean axial stress $` \bar{S}_s(t) `$ in the specimen is given by:
$` \bar{S}_s(t) = \frac{(P_1 + P_2)}{2} \left( \frac{1}{A_s} \right) `$
- Here, $` A_s `$ represents the initial cross-sectional area of the specimen.
5. **Stress-Strain Relationship:**
- Assuming stress equilibrium, uniaxial stress conditions in the specimen, and one-dimensional elastic stress wave propagation without dispersion in the bars, the nominal strain rate $` \dot{e}_s `$, nominal strain $` e_s `$, and nominal stress $` S_s `$ in the specimen are estimated using:
- $` \dot{e}_s(t) = \frac{2c_b}{l_0} \varepsilon_R(t) `$
- $` e_s(t) = \int_0^t \dot{e}_s(\tau) d\tau `$
- $` S_s(t) = \frac{E_b A_b}{A_s} \varepsilon_T(t) `$
6. **True Stress-Strain:**
- True strain $\varepsilon_s(t)$ in the specimen is given by:
- $\varepsilon_s(t) = -\ln(1 - e_s(t))$
Here, $e_s(t)$ represents the engineering strain in the specimen.
- The true strain rate $\dot{\varepsilon}_s(t)$ in the specimen is calculated as:
- $\dot{\varepsilon}_s(t) = \frac{\dot{e}_s(t)}{1 - e_s(t)}$
- The true stress $\sigma_s(t)$ in the specimen is obtained as:
- $\sigma_s(t) = S_s(t) \cdot (1 - e_s(t))$
Here, $S_s(t)$ represents the nominal stress in the specimen.
# SHPB Cropping Tool
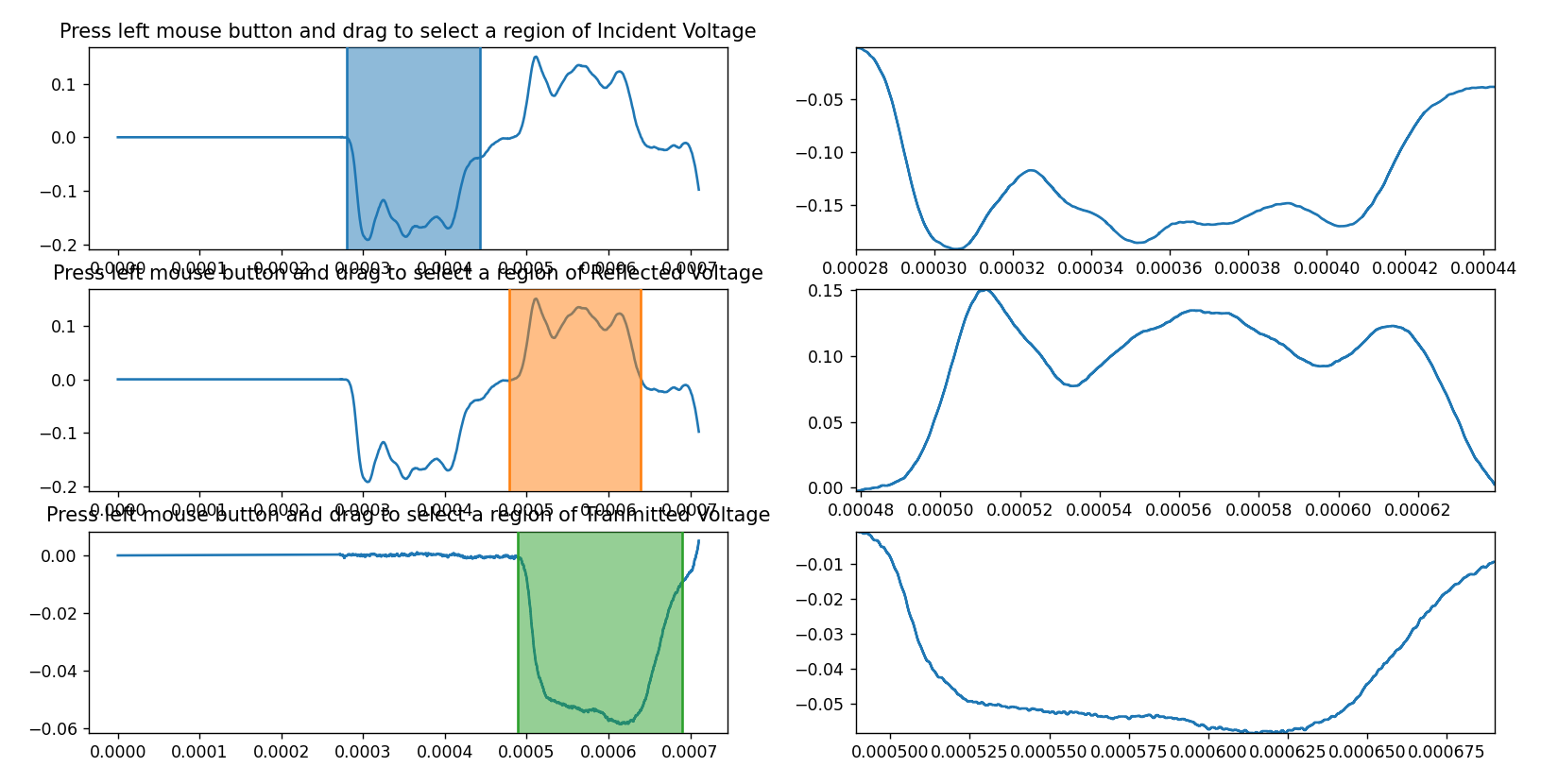
1. **Data Preparation:**
- Prepare your SHPB data in CSV format. Each CSV file should contain three columns: 'Time' and the respective voltage data (e.g., 'Incident' and 'Transmitted').
2. **Launching the Tool:**
- Run the Python script `crop.py`.
- The tool will display a graphical interface with multiple subplots.
3. **Selecting Regions:**
- For Incident Voltage: Click and drag on the subplot (labelled 'Incident Voltage') to select a region of interest.
- For Reflected Voltage: Click and drag on the subplot (labelled 'Reflected Voltage') to select a region of interest.
- For Transmitted Voltage: Click and drag on the subplot (labelled 'Transmitted Voltage') to select a region of interest.
4. **Saving Selected Data:**
- The selected regions will be displayed on their respective subplots.
- The selected data will be saved automatically to CSV files in the same directory as the script.
- Selected Incident Voltage data will be saved to `selected_incident.csv`.
- Selected Reflected Voltage data will be saved to `selected_reflected.csv`.
- Selected Transmitted Voltage data will be saved to `selected_transmitted.csv`.
### Reference
The theory and equations used in this project are based on the following sources:
- Ramesh, K.T. (2008). High Rates and Impact Experiments. In: Sharpe, W. (eds) *Springer Handbook of Experimental Solid Mechanics*. Springer Handbooks. Springer, Boston, MA. [DOI: 10.1007/978-0-387-30877-7_33](https://doi.org/10.1007/978-0-387-30877-7_33)
- Kolsky, H. (1963). Stress Waves in Solids. *United Kingdom: Dover Publications*.
## License
This project is licensed under the Apache-2.0 License. - see the [LICENSE](LICENSE) file for details.