https://github.com/edinburgh-genome-foundry/caravagene
🎨 Python library to plot multi-part genetic constructs
https://github.com/edinburgh-genome-foundry/caravagene
dna-assembly molecular-biology sequence-design synthetic-biology
Last synced: 5 months ago
JSON representation
🎨 Python library to plot multi-part genetic constructs
- Host: GitHub
- URL: https://github.com/edinburgh-genome-foundry/caravagene
- Owner: Edinburgh-Genome-Foundry
- License: mit
- Created: 2017-08-25T17:26:16.000Z (over 7 years ago)
- Default Branch: master
- Last Pushed: 2023-07-21T23:15:30.000Z (almost 2 years ago)
- Last Synced: 2024-11-06T18:52:45.551Z (6 months ago)
- Topics: dna-assembly, molecular-biology, sequence-design, synthetic-biology
- Language: Python
- Homepage: https://edinburgh-genome-foundry.github.io/Caravagene/
- Size: 7.48 MB
- Stars: 25
- Watchers: 6
- Forks: 2
- Open Issues: 0
-
Metadata Files:
- Readme: README.rst
Awesome Lists containing this project
README
.. raw:: html

.. image:: https://github.com/Edinburgh-Genome-Foundry/Caravagene/actions/workflows/build.yml/badge.svg
:target: https://github.com/Edinburgh-Genome-Foundry/Caravagene/actions/workflows/build.yml
:alt: GitHub CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/Caravagene/badge.svg?branch=master
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/Caravagene?branch=master
Caravagene (full documentation `here `_) is a Python library to plot schemas of DNA constructs from a list of parts:
.. code:: python
from caravagene import Part, Construct, ConstructList
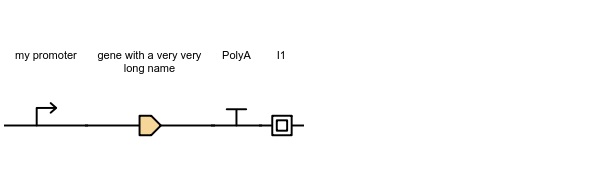
constructs = ConstructList([Construct([
Part('promoter', label='my promoter'),
Part('CDS', label='gene with a very very long name'),
Part('terminator', label='PolyA'),
Part('insulator', label='I1')
])])
constructs.to_image('construct.jpeg')
.. raw:: html
Here is another example producing `this PDF `_
showing multiple constructs:
.. code:: python
from caravagene import Part, Construct, ConstructList
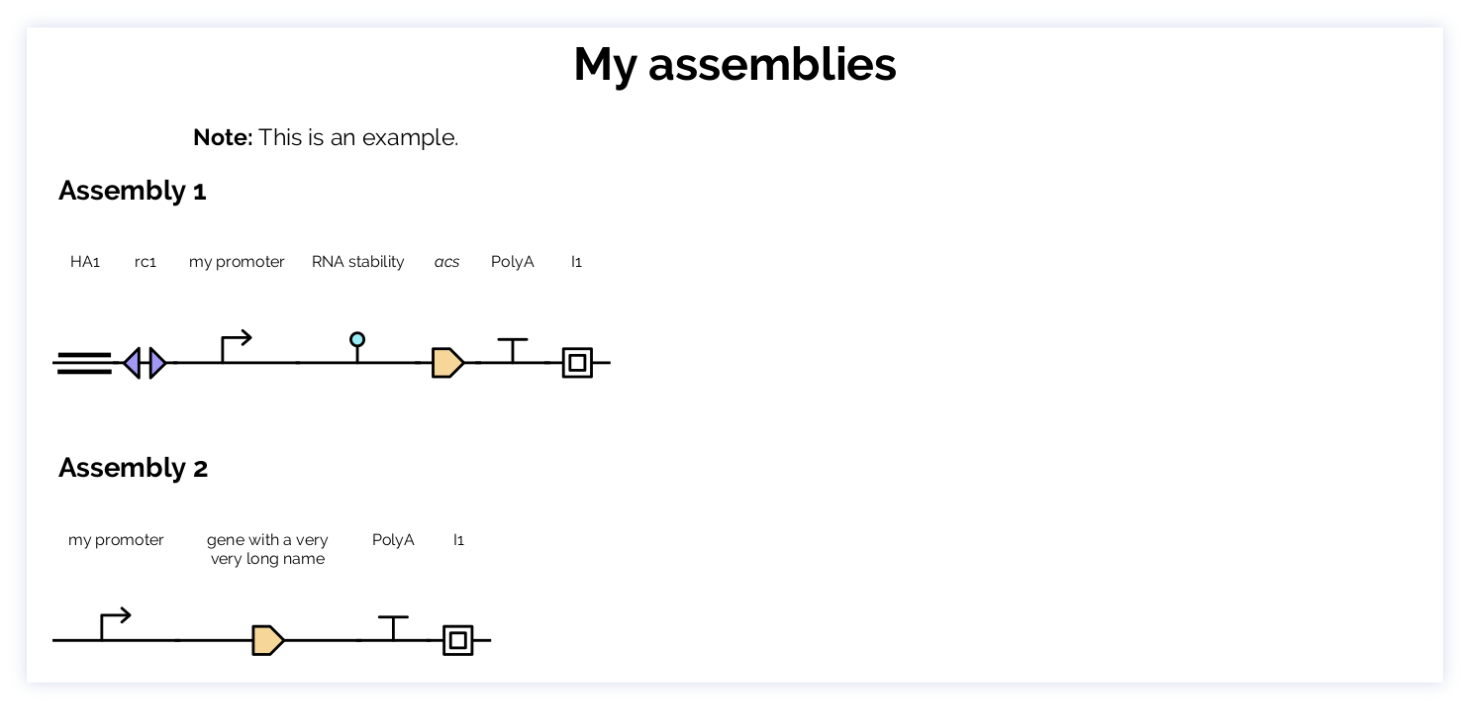
my_constructs = ConstructList(
title="My constructs",
constructs=[
Construct(name="ASM1", parts=[
Part(label='HA1', category='homology-arm'),
Part(label='rc1', category='recombinase-recognition-sequence'),
Part(label='my promoter', category='promoter'),
Part(label='RNA stability', category='rna-stability-sequence'),
Part(label='acs', category='CDS'),
Part(label='PolyA', category='terminator'),
Part(label='I1', category='insulator'),
]),
Construct(name="ASM2", parts=[
Part(label='my promoter', category='promoter'),
Part(label='gene with a very very long name', category='CDS'),
Part(label='PolyA', category='terminator'),
Part(label='I1', category='insulator')
])
]
)
my_constructs.to_pdf('multiconstruct.pdf')
.. raw:: html
Note that ``ConstructList`` can be supplied with a font. It is also possible to extend Caravagene to support other categories/symbols, as follows:
.. code:: python
from caravagene import SYMBOL_FILES
SYMBOL_FILES['my-new-category'] = 'path/to/some/symbol.svg'
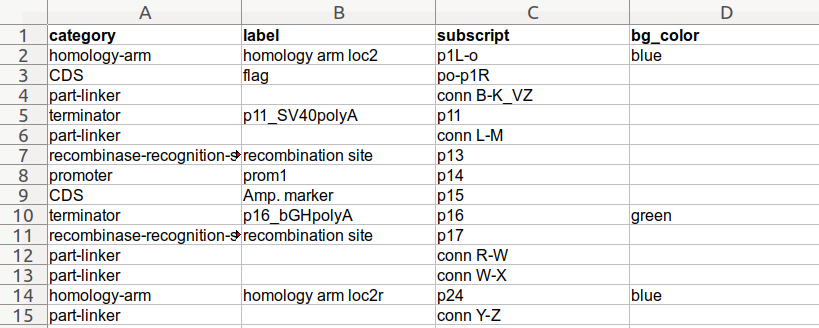
Finally, here is an example using an Excel spreadsheet:
**Spreadsheet:**
.. raw:: html
**Python code:**
.. code:: python
from caravagene import ConstructList
my_constructs = ConstructList("my_spreadsheet.xlsx")
my_constructs.to_pdf('my_schemas.pdf')
or **command-line (one-time use):**
.. code:: shell
caravagene my_spreadsheet.xlsx my_schemas.pdf
or **command-line (re-render when the spreadsheet changes on disk):**
.. code:: shell
caravagene my_spreadsheet.xlsx my_schemas.pdf --watch
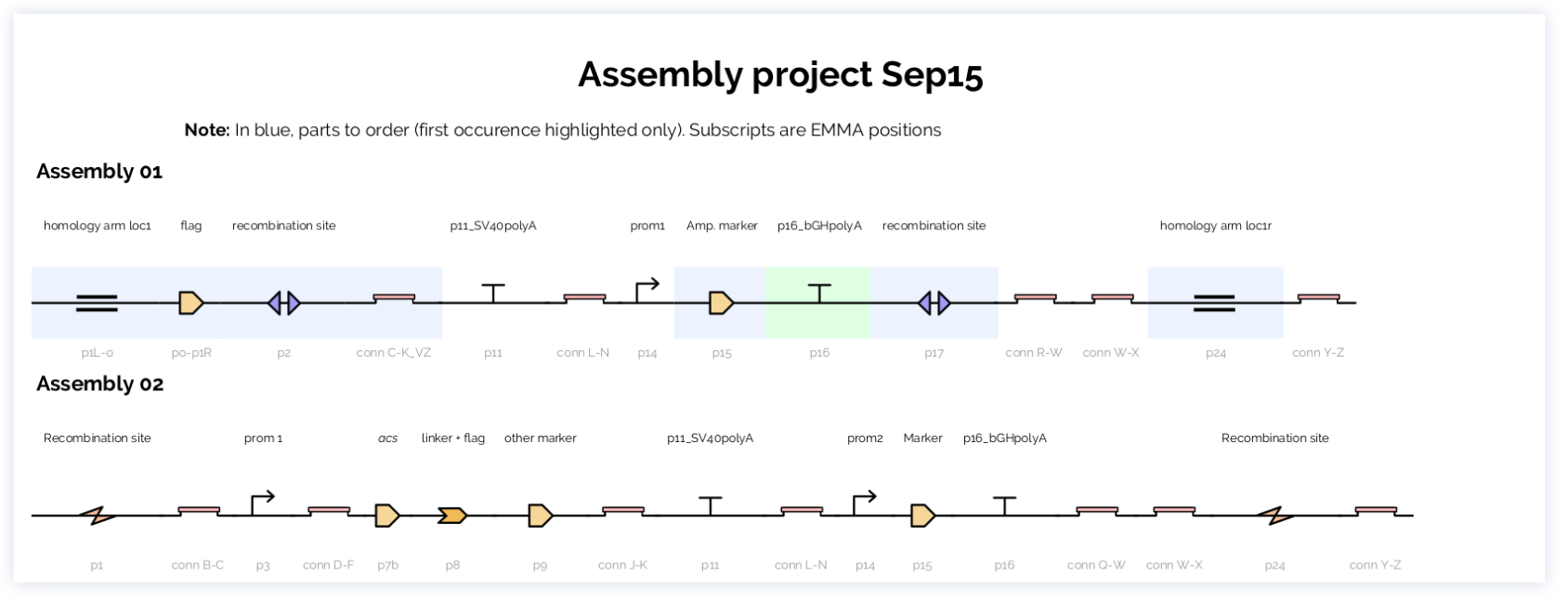
**Output:**
.. raw:: html
Installation
-------------
Caravagene requires `WkHTMLtoPDF `_ to be installed. On Ubuntu, install it with:
.. code:: shell
(sudo) apt-get install wkhtmltopdf
You can install caravagene through PIP:
.. code:: shell
pip install caravagene
Alternatively, you can unzip the source code in a folder and type:
.. code:: shell
sudo python setup.py install
Licence
-------
Caravagene is an open-source software originally written at the `Edinburgh Genome Foundry
`_ by `Zulko `_
and `released on Github `_ under the MIT licence (Copyright 2017 Edinburgh Genome Foundry).
Everyone is welcome to contribute!