https://github.com/edinburgh-genome-foundry/crazydoc
Read DNA sequences from colourful Microsoft Word documents
https://github.com/edinburgh-genome-foundry/crazydoc
bioinformatics computer-aided-design dna-sequences molecular-biology synthetic-biology
Last synced: 3 months ago
JSON representation
Read DNA sequences from colourful Microsoft Word documents
- Host: GitHub
- URL: https://github.com/edinburgh-genome-foundry/crazydoc
- Owner: Edinburgh-Genome-Foundry
- License: mit
- Created: 2018-02-02T23:07:51.000Z (over 7 years ago)
- Default Branch: master
- Last Pushed: 2025-03-31T11:53:47.000Z (7 months ago)
- Last Synced: 2025-04-08T22:16:06.686Z (6 months ago)
- Topics: bioinformatics, computer-aided-design, dna-sequences, molecular-biology, synthetic-biology
- Language: Python
- Homepage: https://edinburgh-genome-foundry.github.io/crazydoc/
- Size: 5.43 MB
- Stars: 32
- Watchers: 5
- Forks: 3
- Open Issues: 0
-
Metadata Files:
- Readme: README.rst
- License: LICENSE
Awesome Lists containing this project
README
.. raw:: html
.. image:: https://github.com/Edinburgh-Genome-Foundry/crazydoc/actions/workflows/build.yml/badge.svg
:target: https://github.com/Edinburgh-Genome-Foundry/crazydoc/actions/workflows/build.yml
:alt: GitHub CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/crazydoc/badge.svg?branch=master
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/crazydoc?branch=master
Crazydoc is a Python library to parse one of the most common DNA representation formats: the joyfully coloured and stylishly annotated MS Word document.
.. raw:: html

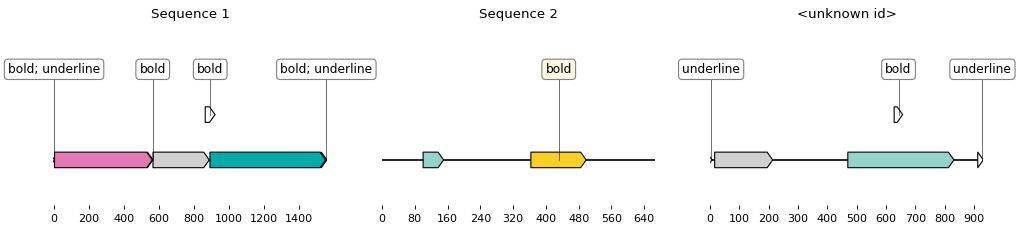
Crazydoc returns Biopython records of the sequences contained in an MS Word document, with record features corresponding to the various sequence highlightings (background color, boldness, italics, case change, etc.). The records can saved as GenBanks or easily plotted.
.. raw:: html
**Motivation**
While other standards such as FASTA or Genbank are better supported by modern sequence editors, none enjoys the same popularity among molecular biologist as MS Word's ``.docx`` format, which is limited only by the sophistication and creativity of the user.
Relying on a loose syntax and unclear specifications, this format has however suffered from a lack of support in the developers community and is generally incompatible with mainstream software pipelines. This library allows to convert MS Word DNA sequences to more computing friendly formats: Biopython records, FASTA, or annotated Genbanks.
Usage
-----
To obtain all sequences contained in a docx as annotated Biopython records (such as `this one `_):
.. code:: python
from crazydoc import CrazydocParser
parser = CrazydocParser(['highlight_color', 'bold', 'underline'])
biopython_records = parser.parse_doc_file("./example.docx")
You can then plot the obtained records:
.. code:: python
from crazydoc import CrazydocSketcher
sketcher = CrazydocSketcher()
for record in biopython_records:
sketch = sketcher.translate_record(record)
ax, _ = sketch.plot()
ax.set_title(record.id)
ax.figure.savefig('%s.png' % record.id)
.. raw:: html

To write the sequences down as Genbank records, with annotations:
.. code:: python
from crazydoc import records_to_genbank
records_to_genbank(biopython_records)
Note that ``records_to_genbank()`` will truncate the record name to 20 characters,
to fit in the GenBank format. Additionally, slashes (``/``) will be replaced with
hyphens (``-``) in the filenames. To read protein sequences, pass ``is_protein=True``:
.. code:: python
biopython_records = parse_doc_file(protein_path, is_protein=True)
This will return *protein* records, which will be saved with a GenPept extension
(.gp) by ``records_to_genbank(biopython_records, is_protein=True)``,
unless specified otherwise with ``extension=``.
You can also save annotated sequences as colourful Word docs.
``write_crazydoc()`` takes a SeqRecord, the qualifier key to use as a feature name,
and a path to save the document to.
.. code:: python
# Load an annotated sequence with Biopython
from Bio import SeqIO
from crazydoc import write_crazydoc
seq = SeqIO.read("examples/examples_outputs/Sequence 1.gbk", "genbank")
# Most features will already have some name qualifier but you can add your own
for i,f in enumerate(seq.features):
f.qualifiers['product'] = f"feature{i}"
# Save the annotated sequence as a docx
write_crazydoc(seq, 'product', 'test.docx')
Installation
------------
You can install crazydoc through PIP:
.. code::
pip install crazydoc
Alternatively, you can unzip the sources in a folder and type:
.. code::
python setup.py install
License = MIT
-------------
Crazydoc is an open-source software originally written at the `Edinburgh Genome Foundry `_ by `Zulko `_ and `released on Github `_ under the MIT licence (Copyright 2018 Edinburgh Genome Foundry).
Everyone is welcome to contribute!
More biology software
---------------------
.. image:: https://raw.githubusercontent.com/Edinburgh-Genome-Foundry/Edinburgh-Genome-Foundry.github.io/master/static/imgs/logos/egf-codon-horizontal.png
:target: https://edinburgh-genome-foundry.github.io/
Crazydoc is part of the `EGF Codons `_ synthetic biology software suite for DNA design, manufacturing and validation.