https://github.com/edinburgh-genome-foundry/genedom
Batch domestication of genetic parts with Python
https://github.com/edinburgh-genome-foundry/genedom
dna-assembly part-domestication sequence-design synbio synthetic-biology
Last synced: 3 months ago
JSON representation
Batch domestication of genetic parts with Python
- Host: GitHub
- URL: https://github.com/edinburgh-genome-foundry/genedom
- Owner: Edinburgh-Genome-Foundry
- License: mit
- Created: 2018-04-13T16:37:54.000Z (over 7 years ago)
- Default Branch: master
- Last Pushed: 2025-05-12T14:14:27.000Z (6 months ago)
- Last Synced: 2025-07-15T09:03:07.304Z (3 months ago)
- Topics: dna-assembly, part-domestication, sequence-design, synbio, synthetic-biology
- Language: Python
- Homepage: https://edinburgh-genome-foundry.github.io/genedom/
- Size: 2.51 MB
- Stars: 16
- Watchers: 5
- Forks: 1
- Open Issues: 0
-
Metadata Files:
- Readme: README.rst
- License: LICENSE
Awesome Lists containing this project
README
.. raw:: html

.. image:: https://github.com/Edinburgh-Genome-Foundry/genedom/actions/workflows/build.yml/badge.svg
:target: https://github.com/Edinburgh-Genome-Foundry/genedom/actions/workflows/build.yml
:alt: GitHub CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/genedom/badge.svg?branch=master
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/genedom?branch=master
Genedom is a Python package for managing the domestication of genetic parts,
which means modifying their sequences to make them compatible with a given
genetic assembly standard. Genedom binds together a
`sequence optimizer `_,
information on the genetic standard, and a reporting routine to automate the
domestication of large batches in an easy and human-friendly way.
.. raw:: html
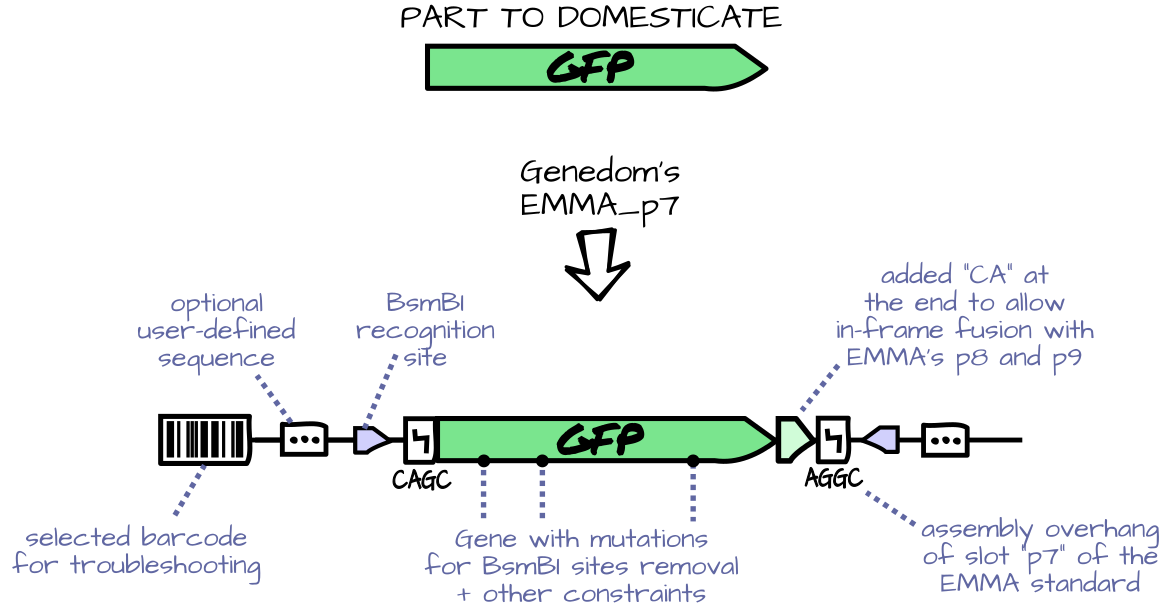
Features include:
- User-defined part domesticators with extra nucleotides optionally added to both ends,
hard constraints on the sequence (such as enforcing absence of a restriction
site) and optimization objectives (such as codon optimization).
- Built-in pre-defined domesticators for popular genetic assembly standards
(well, only EMMA at the moment).
- Generation of barcodes that can be added to the sequence
(but won't be in final constructs). This allows easy verification of a sequence file
or a DNA sample (e.g. in case of label mix-up).
- Routine for batch-domesticating sequences with report generation, including
reports on each sequence optimization, spreadsheets of parts, ready-to-order FASTA
and Genbank files of the parts, and a summary report to quickly verify everything,
with a list of every domesticator used, for traceability.
An example summary report:
.. raw:: html
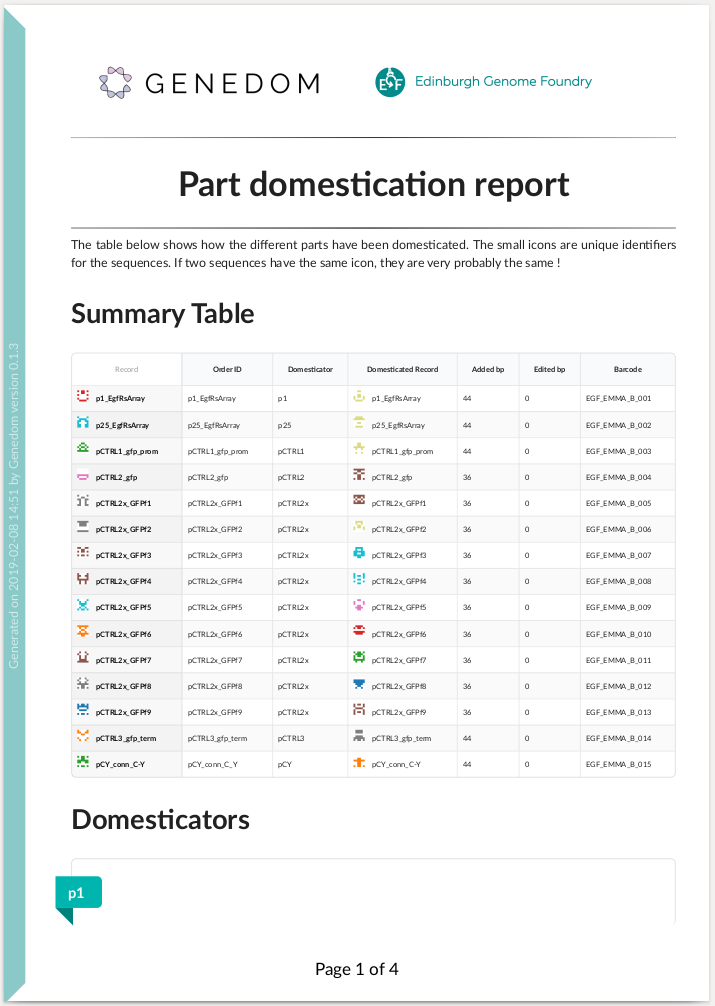
You can also use Genedom online via EGF's `Domesticate Part Batches web app `_.
Usage examples
--------------
Simple domestication of one part
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
.. code:: python
from genedom import (GoldenGateDomesticator, random_dna_sequence,
write_record)
sequence = random_dna_sequence(2000, seed=123)
domesticator = GoldenGateDomesticator("ATTC", "ATCG", enzyme='BsmBI')
domestication_results = domesticator.domesticate(sequence, edit=True)
print (domestication_results.summary())
write_record(domestication_results.record_after, 'domesticated.gb')
Generating a collection of 20bp barcodes
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
(see docs for more options)
.. code:: python
from genedom import BarcodesCollection
barcodes_collection = BarcodesCollection.from_specs(
n_barcodes=96, barcode_length=20,
forbidden_enzymes=('BsaI', 'BsmBI', 'BbsI'))
barcodes_collection.to_fasta('example_barcodes_collection.fa')
Domesticating a batch of parts with PDF report
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
.. code:: python
from genedom import BUILTIN_STANDARDS, load_record, batch_domestication
records = [
load_record(filepath, name=filename)
for filepath in records_filepaths
]
barcodes_collection = BarcodesCollection.from_specs(n_barcodes=10)
batch_domestication(records, 'domestication_report.zip',
barcodes=barcodes, # optional
standard=BUILTIN_STANDARDS.EMMA)
Installation
------------
You can install Genedom through PIP:
.. code:: shell
pip install genedom
License = MIT
-------------
Genedom is a free/libre and open-source software
`released on Github `_ under
the MIT license (Copyright 2018 Edinburgh Genome Foundry, University of Edinburgh).
It was originally written by `Zulko `_ and is currently
being developed by `Peter Vegh `_.
Everyone is welcome to `contribute `_!