https://github.com/edinburgh-genome-foundry/kappagate
:crystal_ball: Predict DNA assembly clone validity rates - powered by Kappa
https://github.com/edinburgh-genome-foundry/kappagate
biological-simulations dna-assembly kappa-language lab-automation sequence-design synthetic-biology
Last synced: about 1 month ago
JSON representation
:crystal_ball: Predict DNA assembly clone validity rates - powered by Kappa
- Host: GitHub
- URL: https://github.com/edinburgh-genome-foundry/kappagate
- Owner: Edinburgh-Genome-Foundry
- License: other
- Created: 2018-12-21T19:13:59.000Z (over 6 years ago)
- Default Branch: master
- Last Pushed: 2020-07-24T16:35:08.000Z (almost 5 years ago)
- Last Synced: 2025-03-15T23:41:27.628Z (about 1 month ago)
- Topics: biological-simulations, dna-assembly, kappa-language, lab-automation, sequence-design, synthetic-biology
- Language: Python
- Homepage:
- Size: 257 KB
- Stars: 9
- Watchers: 3
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.rst
Awesome Lists containing this project
README
.. raw:: html

.. image:: https://travis-ci.org/Edinburgh-Genome-Foundry/kappagate.svg?branch=master
:target: https://travis-ci.org/Edinburgh-Genome-Foundry/kappagate
:alt: Travis CI build status
.. image:: https://coveralls.io/repos/github/Edinburgh-Genome-Foundry/kappagate/badge.svg?branch=master
:target: https://coveralls.io/github/Edinburgh-Genome-Foundry/kappagate?branch=master
Kappagate is a Python library to predict the percentage of good clones (carrying
a correct version of the desired assembly) when assembling DNA with a method
relying on 4bp overhangs (e.g. Golden Gate assembly, OGAB, etc.).
Using Kappagate, you can get an estimation of how difficult the
assembly will be, and how many clones should be tested to find a correct one.
Kappagate uses the exhaustive relative overhang affinity tables provided
in Potapov et. al. 2018 (ACS Syn. Bio.). In this publication the authors show
that the proportion of valid clones rates can be predicted using focused
in-vitro experiments focused on the overhangs present in the assembly.
Kappagate attempts to predict clone validity rates without any overhang-subset-specific
experiment, using computer simulations instead. It simulates the temporal evolution
of the DNA fragments ligation reaction using the Kappa biological modeling system.
At the end of the cloning simulation, Kappagate returns the ratio between "good"
constructs (with all expected parts in the right order) and bad circular assembly-forming
constructs (which may produce bad clones after transformation and plating).
This is an experimental piece of software, useful to us, but coming with no warranty.
Examples
--------
.. code:: python
from kappagate import predict_assembly_accuracy, overhangs_list_to_slots
# FIRST TEST ON 12 WELL-DESIGNED OVERHANGS
overhangs= ['GGAG', 'GGCA', 'TCGC', 'CAGT', 'TCCA',
'GAAT', 'AGTA', 'TCTT', 'CAAA', 'GCAC',
'AACG', 'GTCT', 'CCAT']
slots = overhangs_list_to_slots(overhangs)
predicted_rate, _, _ = predict_assembly_accuracy(slots)
print (predicted_rate)
# >>> 0.987
This means that 98.7% of clones will carry a valid assembly. It is really
not far from the experimental observation in Potapov et al., which was
99.2% +- 0.6% (1 std).
Let's have a look at a few more sets:
.. code:: python
overhangs = ['GGAG', 'GATA', 'GGCA', 'GGTC', 'TCGC',
'GAGG', 'CAGT', 'GTAA', 'TCCA', 'CACA',
'GAAT', 'ATAG', 'AGTA', 'ATCA', 'TCTT',
'AGGT', 'CAAA', 'AAGC', 'GCAC', 'CAAC',
'AACG', 'CGAA', 'GTCT', 'TCAG', 'CCAT']
slots = overhangs_list_to_slots(overhangs)
predicted_rate, _, _ = predict_assembly_accuracy(slots)
print (predicted_rate)
# >>> 0.846
# In Potapov 2018: 84% +/- 5%
.. code:: python
overhangs= ['GGAG', 'GGTC', 'AGCA', 'CAGT', 'GGTA',
'GAAT', 'GGTT', 'TCTT', 'GGTG', 'GCAC',
'AGCG', 'GTCT', 'CCAT']
slots = overhangs_list_to_slots(overhangs)
predicted_rate, _, _ = predict_assembly_accuracy(slots)
print (predicted_rate)
# >>> 0.33
# In Potapov 2018: 45% +/- 5%
Moar examples !!
----------------
Plotting interactions
~~~~~~~~~~~~~~~~~~~~~
To plot the parts circularly with their interaction:
.. code:: python
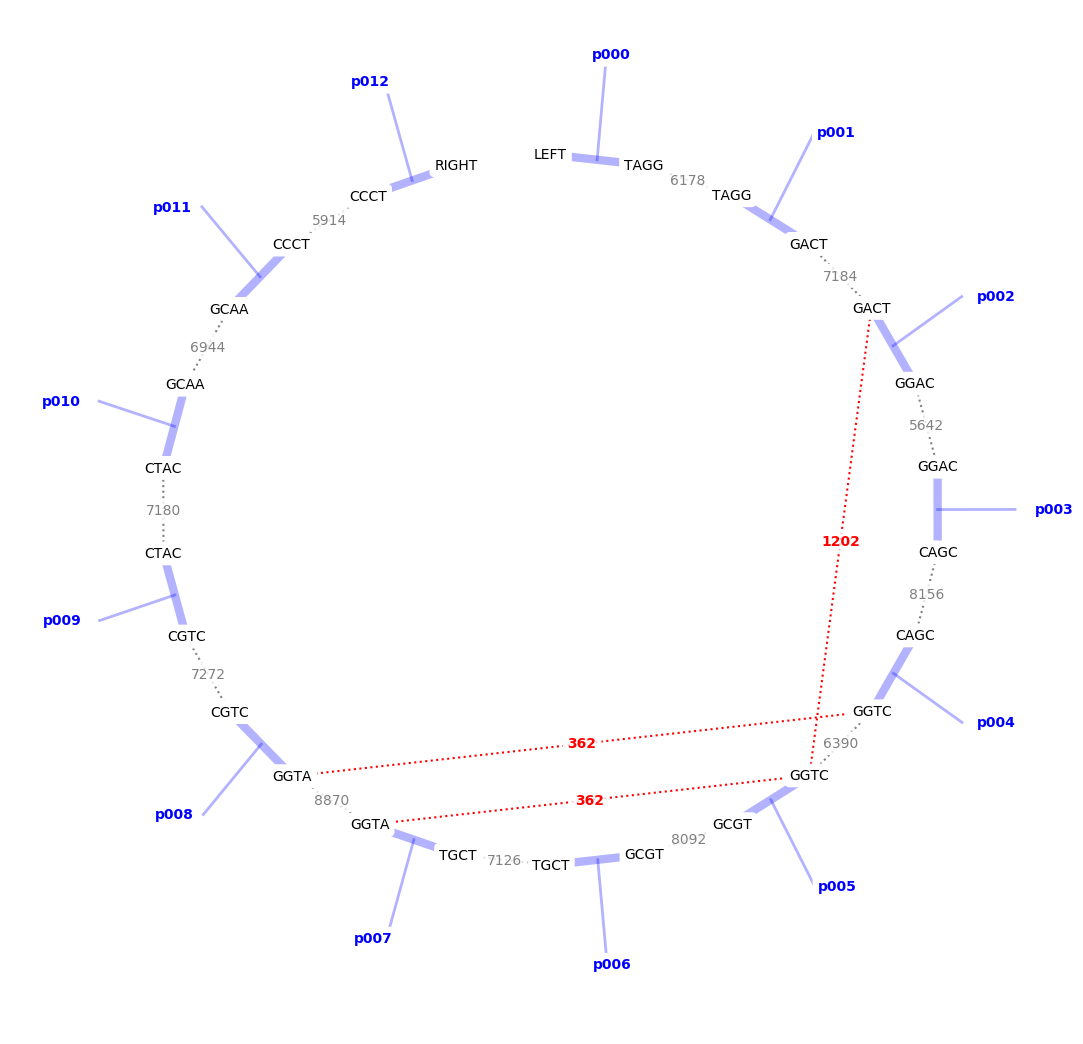
from kappagate import overhangs_list_to_slots, plot_circular_interactions
overhangs = ['TAGG', 'GACT', 'GGAC', 'CAGC',
'GGTC', 'GCGT', 'TGCT', 'GGTA',
'CGTC', 'CTAC', 'GCAA', 'CCCT']
slots = overhangs_list_to_slots(overhangs)
ax = plot_circular_interactions(
slots, annealing_data=('25C', '01h'), rate_limit=200)
ax.figure.savefig("test.png", bbox_inches='tight')
The unwanted overhang interactions appear in red in the resulting figure:
.. raw:: html
Colony picking statistics
~~~~~~~~~~~~~~~~~~~~~~~~~
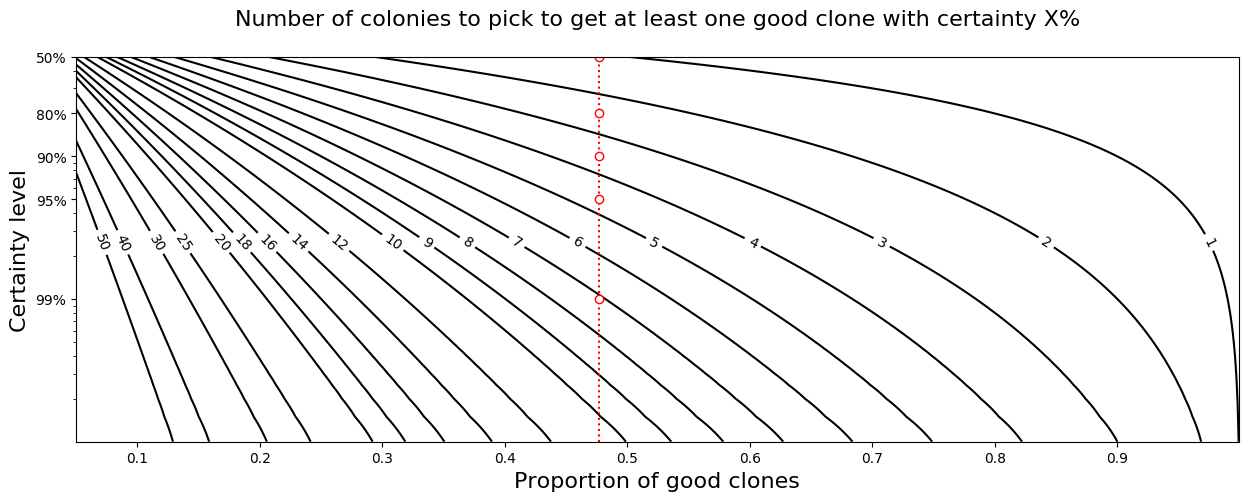
To convert the predicted success rate into decisions regarding how many colonies
to pick, and when to stop picking colonies:
.. code:: python
from kappagate import (overhangs_list_to_slots, predict_assembly_accuracy,
plot_colony_picking_graph, success_rate_facts)
overhangs = ['TAGG', 'GACT', 'GGAC', 'CAGC',
'GGTC', 'GCGT', 'TGCT', 'GGTA',
'CGTC', 'CTAC', 'GCAA', 'CCCT']
slots = overhangs_list_to_slots(overhangs)
predicted_rate, _, _ = predict_assembly_accuracy(slots)
ax = plot_colony_picking_graph(success_rate=predicted_rate)
ax.figure.savefig("success_rate_facts.png", bbox_inches='tight')
print (success_rate_facts(predicted_rate, plain_text=True))
Result:
.. code:: raw
The valid colony rate is 47.7%. Expect 1.9 clones in average
until success. Pick 5 clones or more for 95% chances of at
least one success. If no success after 8 clones, there is
likely another problem (p-value=0.01).
.. raw:: html
Installation
-------------
You can install kappagate through PIP
.. code::
sudo pip install kappagate
Alternatively, you can unzip the sources in a folder and type
.. code::
sudo python setup.py install
License = MIT
--------------
Kappagate is an open-source software originally written at the `Edinburgh Genome Foundry `_ by `Zulko `_ and `released on Github `_ under the MIT licence (Copyright 2018 Edinburgh Genome Foundry).
Everyone is welcome to contribute !
More biology software
---------------------
.. image:: https://raw.githubusercontent.com/Edinburgh-Genome-Foundry/Edinburgh-Genome-Foundry.github.io/master/static/imgs/logos/egf-codon-horizontal.png
:target: https://edinburgh-genome-foundry.github.io/
Kappagate is part of the `EGF Codons `_ synthetic biology software suite for DNA design, manufacturing and validation.