https://github.com/evamaxfield/papers-without-code
A package (and website) to automatically attempt to find the code associated with a paper.
https://github.com/evamaxfield/papers-without-code
Last synced: 8 months ago
JSON representation
A package (and website) to automatically attempt to find the code associated with a paper.
- Host: GitHub
- URL: https://github.com/evamaxfield/papers-without-code
- Owner: evamaxfield
- License: mit
- Created: 2022-11-23T19:06:53.000Z (over 3 years ago)
- Default Branch: main
- Last Pushed: 2025-04-01T08:07:50.000Z (11 months ago)
- Last Synced: 2025-06-23T11:07:55.486Z (8 months ago)
- Language: Jupyter Notebook
- Homepage: https://paperswithoutcode.org
- Size: 12.6 MB
- Stars: 4
- Watchers: 1
- Forks: 0
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- Contributing: CONTRIBUTING.md
- License: LICENSE
- Code of conduct: CODE_OF_CONDUCT.md
Awesome Lists containing this project
README
# papers-without-code
[](https://github.com/evamaxfield/papers-without-code/actions)
[](https://evamaxfield.github.io/papers-without-code)
A Python package ([and website](https://paperswithoutcode.org)) to automatically attempt to find GitHub
repositories that are similar to academic papers.
[](https://paperswithoutcode.org)
---
## Installation
**Stable Release:** `pip install papers-without-code`
**Development Head:** `pip install git+https://github.com/evamaxfield/papers-without-code.git`
## Usage
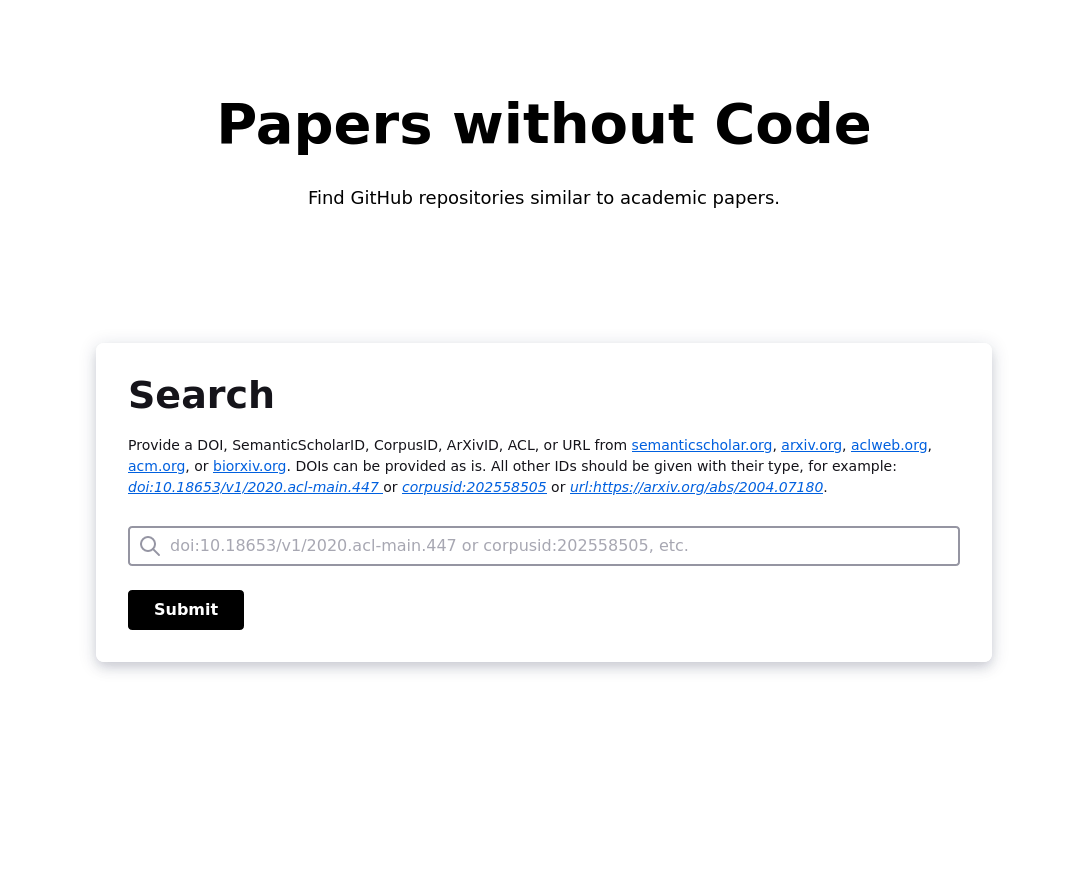
Provide a DOI, SemanticScholarID, CorpusID, ArXivID, ACL,
or URL from semanticscholar.org, arxiv.org, aclweb.org,
acm.org, or biorxiv.org. DOIs can be provided as is.
All other IDs should be given with their type, for example:
`doi:10.18653/v1/2020.acl-main.447`
or `CorpusID:202558505` or `url:https://arxiv.org/abs/2004.07180`.
### CLI
```bash
pip install papers-without-code
pwoc query
# or pwoc path/to/file.pdf
```
### Python
```python
from papers_without_code import search_for_repos
search_for_repos("query")
# search_for_repos("path/to/file.pdf")
```
⚠️ Prior to using PWOC with a PDF you must be logged in to Docker CLI via `docker login`
because we automatically fetch, spin up, and tear down containers for processing. ⚠️
## How it Works
In short, we pass the query on to the Semantic Scholar search API
which provides us basic details about the paper. We use
a prompted gpt-3.5-turbo with langchain to extract keywords from the
title and abstract. We then make multiple threaded requests to GitHub's API
for repositories which match the keywords. Once we have all the possible repositories
back, we rank them by similarity between the repository's README and the paper's
abstract (or if not available, it's title).
When using Papers without Code locally and providing a filepath, the only change to
this workflow, is paper details gathering. When local and providing a filepath,
we use [GROBID](https://github.com/kermitt2/grobid) to extract the
title, abstract, and author list.
## Documentation
For full package documentation please visit [evamaxfield.github.io/papers-without-code](https://evamaxfield.github.io/papers-without-code).
[Exploratory data analysis of the dataset used for testing](https://evamaxfield.github.io/papers-without-code/eda.html)
## Development
See [CONTRIBUTING.md](CONTRIBUTING.md) for information related to developing the code.
**MIT License**