Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/faisalthaheem/simanno
Simple Image Annotation (simanno) Tool for Machine Learning Projects
https://github.com/faisalthaheem/simanno
annotation-tool image-processing machine-learning tool
Last synced: 3 months ago
JSON representation
Simple Image Annotation (simanno) Tool for Machine Learning Projects
- Host: GitHub
- URL: https://github.com/faisalthaheem/simanno
- Owner: faisalthaheem
- License: apache-2.0
- Created: 2018-07-03T13:05:23.000Z (over 6 years ago)
- Default Branch: master
- Last Pushed: 2022-02-07T05:46:26.000Z (about 3 years ago)
- Last Synced: 2024-08-02T15:25:51.247Z (7 months ago)
- Topics: annotation-tool, image-processing, machine-learning, tool
- Size: 10.7 MB
- Stars: 5
- Watchers: 5
- Forks: 1
- Open Issues: 3
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
|Repository|Last commit|Docker CI Health|Docker pulls|
|---|---|---|---|
|Scripts||[](https://github.com/faisalthaheem/simanno-scripts/actions/workflows/docker-image.yml)||
|Backend||[](https://github.com/faisalthaheem/simanno-be/actions/workflows/docker-image.yml)||
|Frontend||[](https://github.com/faisalthaheem/simanno-fe/actions/workflows/docker-image.yml)||
# simanno
Simple Image Annotation (SimAnno) Tool for Deep Learning Projects.
SimAnno is a web based image annotation tool that uses sqlite database(s) to maintain annotations for images and consists of a front end which is an app written in Angular 8 and is supported by a Flask based python 3 REST service backend.
This repository has been refactored to serve as the documentation and general information repo, for specific modules please refer to the respective repositories below.
|Module|Repo link|
|---|---|
|Documentation|This repo|
|Backend|https://github.com/faisalthaheem/simanno-be|
|Frontend|https://github.com/faisalthaheem/simanno-fe|
|Scripts|https://github.com/faisalthaheem/simanno-scripts|
At the heart of the project is the great [jquery select areas plugin](https://github.com/360Learning/jquery-select-areas).
The tool makes it easy to keep images on a headless environment and access them from any web browser. Once images have been tagged a training record can be generated to be fed to the learning/evaluating scripts. See [life2tfrec.py](https://github.com/faisalthaheem/deep-learning-scripts/blob/master/dbutils/lite2tfrec.py) for an example of how the databases generated by this tool are being used.
The easiest and straightforward way is to use Docker to run the services, see [Running with Docker](https://github.com/faisalthaheem/simanno/wiki/Running-with-Docker) for more information.
If you would like to run outside of Docker, you will need to install dependencies, please refer to [Setup Guide](https://github.com/faisalthaheem/simanno/wiki/Setup-Guide) to get started.
# Demo
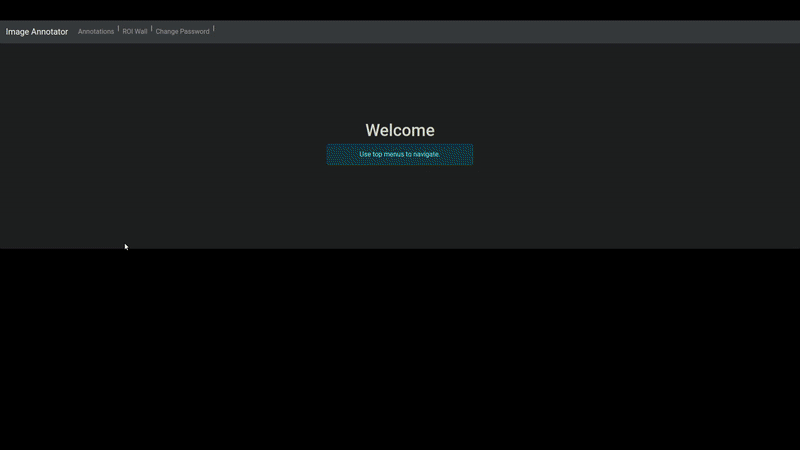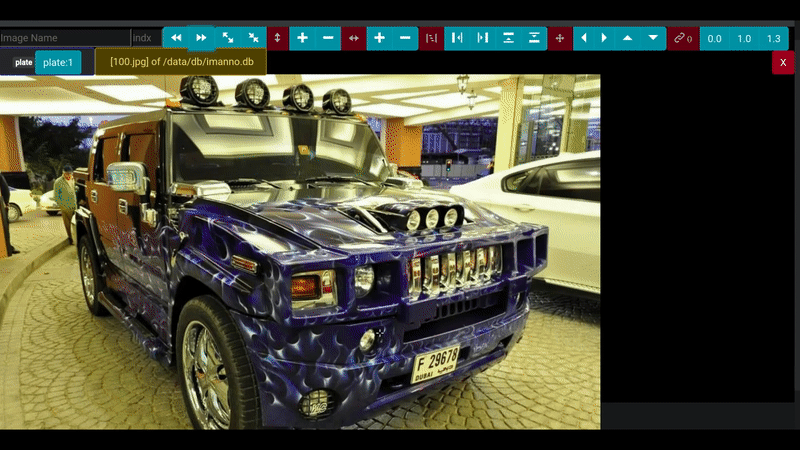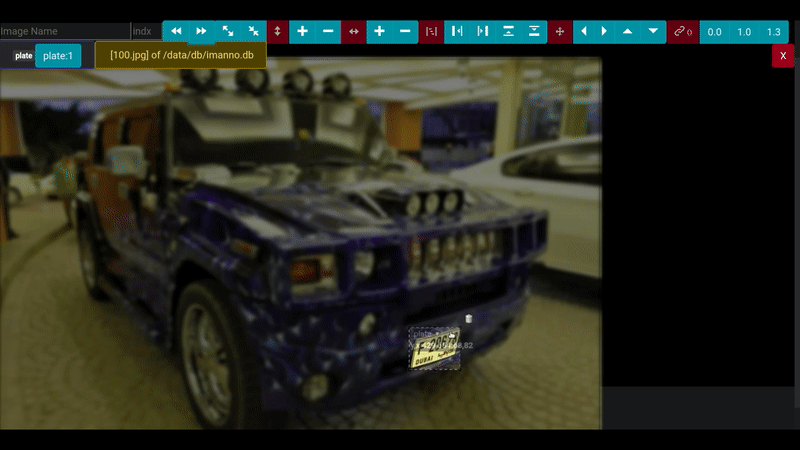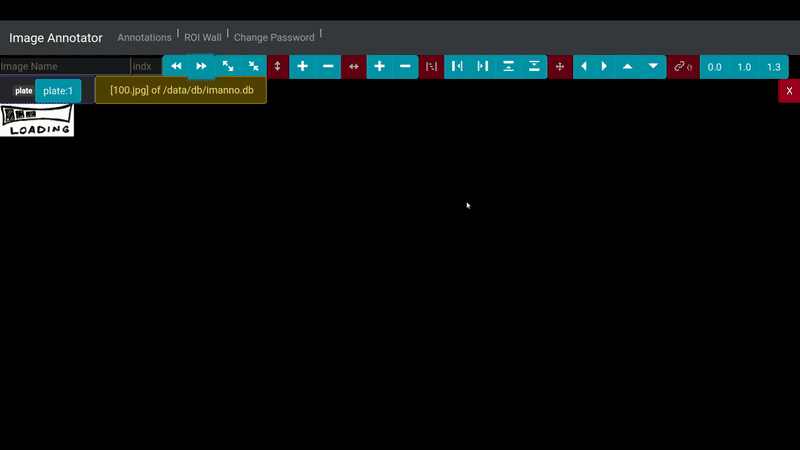
## Annotation Screen
Following screen a sample image with different areas marked with appropriate labels

## ROI (Regions Of Interest) Wall
Following image shows the regions of interest extracted from the available data for a quick review

## Running the demo locally
To launch the stack using the latest containers from main branch, use the following command
```bash
docker-compose -f docker-compose.yml up -d
```
If you have cloned the repositories and are developing, you can use the following command to launch the stack in development mode
```bash
docker-compose -f docker-compose.dev.yml up -d
```
# Issues and Feedback
Please use the issues link on the top to report any issues/feedback/suggestions.
# Scripts
There are few useful scripts that faciliate data import and export to/from simanno. The following section describes these briefly.
## code/import-cat-from-coco.py
> Using this script you can extract a category of images from the COCO dataset.
The scripts expects the following arguments
The comand expects the following arguments
```bash
usage: import-cat-from-coco.py [-h] -c CATEGORY -t TYPE -li LBLID -af ANNOTATIONFILE -sp SRCPATH -dp DESTPATH
optional arguments:
-h, --help show this help message and exit
-c CATEGORY, --category CATEGORY
The category to extract.
-t TYPE, --type TYPE Type to assign to images, accepted values are train or val, default is train.
-li LBLID, --lblid LBLID
The label id to be used for this category.
-af ANNOTATIONFILE, --annotationfile ANNOTATIONFILE
Path to annotation file.
-sp SRCPATH, --srcpath SRCPATH
Path of source containing images.
-dp DESTPATH, --destpath DESTPATH
Path to destination to create the db and save images to.
```
For instance, the following command extracts all images belonging to the "cars" category and moves them to a destination folder while also creating a stand alone sqlite db that works with simanno.
```bash
python3 import-cat-from-coco.py \
-t val -c car -li 1 \
-af $COCO_PATH/annotations/instances_val2017.json
-dp $CARS_FROM_COCO_PATH
-sp $COCO_PATH/val2017/
```
Where, the parameters are
|Parameter|Valid values|Description|
|---|---|---|
|-t |val or train| Type of dataset that is being created for simanno. Influences the names of db file and destination folder.|
|-c |string| Name of category that should exist in coco dataset.|
|-li|integer| The label index to be assigned when records are added to db. Label text is taken from the -c parameter.|
|-af|path| Path to the coco annotations file.|
|-dp|path| Path to the destination folder which will contain a database file and a folder containing images copied from the coco dataset.|
|-sp|path| Path to the source folder containing coco images to be copied from into the destination folder.|
Alternatively you can use the docker-container version to run this script with the following command
```bash
docker run --rm -it -u $UID -v $PWD:/datasets faisalthaheem/simanno-scripts:main "/usr/local/bin/python3.8 import-cat-from-coco.py -t val -c car -li 1 -af /datasets/coco/annotations/instances_val2017.json -dp /datasets/cars_from_coco -sp /datasets/coco/val2017/"
```
```bash
docker run --rm -it -u $UID -v $PWD:/datasets faisalthaheem/simanno-scripts:main "/usr/local/bin/python3.8 import-cat-from-coco.py -t train -c car -li 1 -af /datasets/coco/annotations/instances_train2017.json -dp /datasets/cars_from_coco -sp /datasets/coco/train2017/"
```
> Notice "/usr/local/bin/python3.8" which is important in the image
## code/mergedbs.py
> For merging multiple simanno datasets into a single set for training and validation.
This script uses a yaml configuration file as input and provides remapping of the source labels in addition to merging annotations db table and files.
As output train and val folders containing images and train and val dbs are produced.
If the destination dbs exist then they are deleted before re-generation, files are only copied again if missing in destination.
For an example of a sample merge config file please take a look at
```bash
conf/scripts/mergedbs.sample.yaml
```
The comand expects the following arguments
```bash
usage: mergedbs.py [-h] -c CONFIGFILE
optional arguments:
-h, --help show this help message and exit
-c CONFIGFILE, --configfile CONFIGFILE
Path to the merge config file.
```
To execute, use the following command
```bash
python3 mergedbs.py -c mergedbs.yaml
```
Or, use the docker variant as given below, assuming you're in the folder containing all your datasets
```bash
docker run --rm -it -u $UID -v $PWD:/datasets faisalthaheem/simanno-scripts:main "/usr/local/bin/python3.8 /simanno/scripts/mergedbs.py -c /datasets/mergedbs.yaml"
```
## code/splitdb.py
> For splitting a common db into two given input a folder containing images.
Given there is a simanno database that contains annotation of images contained in several folders, this script allows to split this database into several independent files thus allowing for better management of the training data.
To execute the script directly, a command similar to following can be used
```bash
python3 splitdb.py -r $PLATE_DETECTION_PATH/train -o $PLATE_DETECTION_PATH/train.db -i $PLATE_DETECTION_PATH/train_and_val.d
```
Or, if using the docker image, then following command accomplishes the same result
```bash
docker run --rm -it -u $UID -v $PWD:/datasets faisalthaheem/simanno-scripts:main "/usr/local/bin/python3.8 /simanno/scripts/splitdb.py -r /datasets/plate_detection/train -o /datasets/plate_detection/train.db -i /datasets/plate_detection/train_and_val.db"
```
The comand expects the following arguments
```bash
usage: splitdb.py [-h] -r REF -o OUT -i IN
optional arguments:
-h, --help show this help message and exit
-r REF, --ref REF Path to the folder containing ref images.
-o OUT, --out OUT Path to the destination db which will be created bearing contents related to ref folder images.
-i IN, --in IN Path to the source db containing information on ref images.
```