https://github.com/firstmover/cr-seg
Source code for "Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series" paper.
https://github.com/firstmover/cr-seg
consistency-regularization mri registration segmentation semi-supervised-learning
Last synced: 4 months ago
JSON representation
Source code for "Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series" paper.
- Host: GitHub
- URL: https://github.com/firstmover/cr-seg
- Owner: firstmover
- License: mit
- Created: 2023-10-05T18:10:34.000Z (almost 2 years ago)
- Default Branch: main
- Last Pushed: 2023-10-16T03:12:36.000Z (over 1 year ago)
- Last Synced: 2025-01-24T15:43:12.390Z (6 months ago)
- Topics: consistency-regularization, mri, registration, segmentation, semi-supervised-learning
- Language: Python
- Homepage:
- Size: 72.3 KB
- Stars: 0
- Watchers: 1
- Forks: 1
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
## Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series
This repo includes the code for the paper [Liu et al. 2023](https://arxiv.org/pdf/2310.03870.pdf).

[](https://github.com/firstmover/cr-seg)
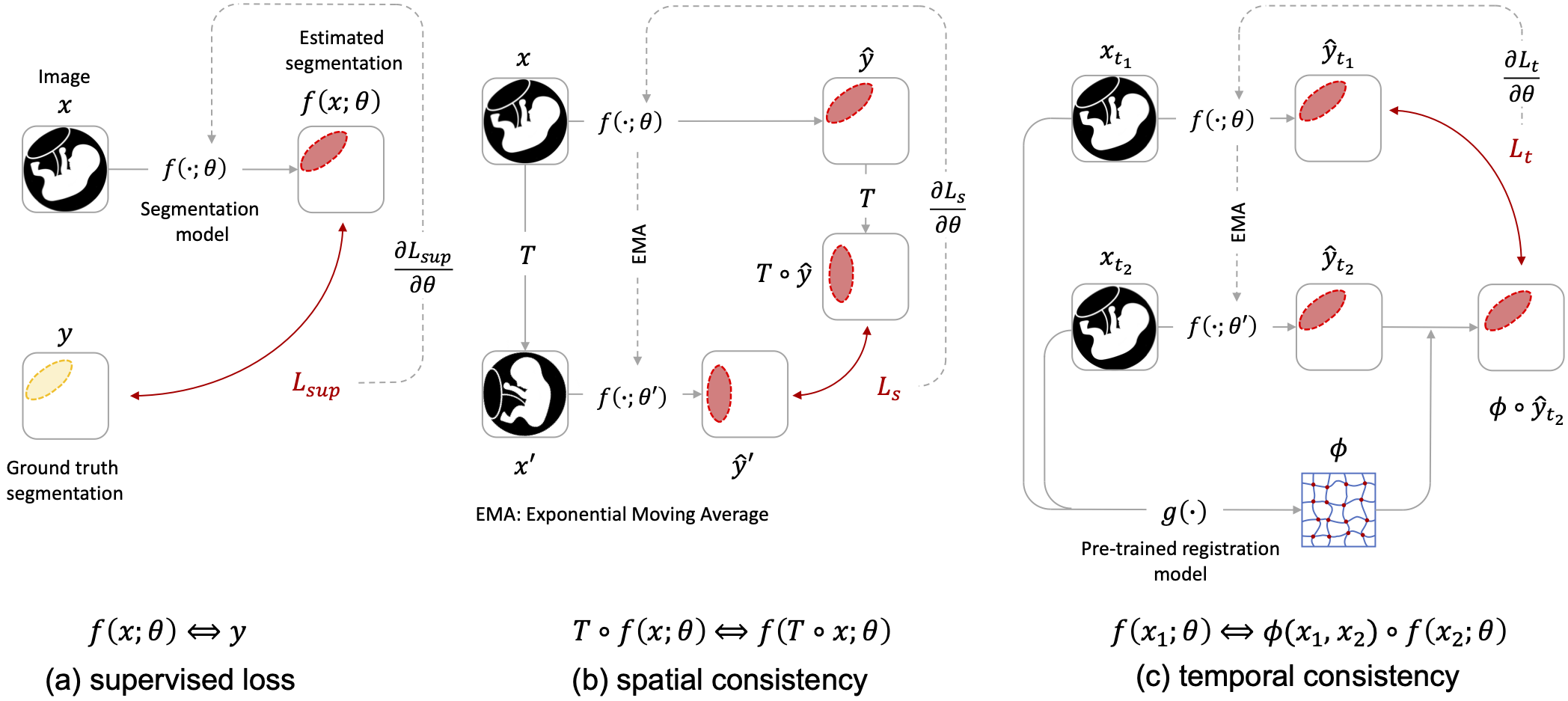
### Environment and dependency
Create a conda environment with:
```bash
conda create --name cr_seg --file requirements.txt
```
Specify your paths to data, cache, and results directories in:
1. `./envs/default`
2. `./configs/segm_release/spatial_temporal_cr.py`
### Results
#### Pre-process of data
```bash
srun --partition=gpu \
--job-name=segm \
--gres=gpu:1 \
--ntasks=1 \
--ntasks-per-node=1 \
--cpus-per-task=16 \
--time=24:00:00 \
python scripts/pre_compute_data.py
```
#### Training
pretrain registration models for all cross-validation folds
```bash
python ./scripts/submit_job_registraion.py \
--exp-name regi_release \
--config-name voxelmorph \
--job-name=regi \
--num-gpus-per-node=1 \
--cpus-per-task=20 \
--num-nodes=1 \
--array-parallelism=5
```
Train UNet with spatial and temporal consistency regularization for all cross-validation folds
```bash
python ./scripts/submit_job_segmentation.py \
--task-mode train \
--exp-name segm_release \
--config-name spatial_temporal_cr \
--lambda-list '0.001' \
--lambda-t-list '0.001' \
--job-name segm_regi \
--num-gpus-per-node 4 \
--cpus-per-task 8 \
--array-parallelism 5
```
#### Inference and evaluation
Run inference for labeled data for all cross-validation folds
```bash
python ./scripts/submit_job_segmentation.py \
--task-mode inference_labeled \
--exp-name segm_release \
--config-name spatial_temporal_cr \
--lambda-list '0.001' \
--lambda-t-list '0.001' \
--tta \
--tta-cfg-path ./configs/segm_release/_base_/tta_all_crop.py \
--save-data-name-list 'img,pred_seg_map,gt_seg_map' \
--job-name inference \
--partition gpu \
--num-gpus-per-node 1 \
--cpus-per-task 16 \
--array-parallelism 5
```
Run inference for time series data (unlabeled and labeled data) for all cross-validation folds
```bash
python ./scripts/submit_job_segmentation.py \
--task-mode inference_time_series \
--exp-name segm_release \
--config-name spatial_temporal_cr \
--lambda-list '0.001' \
--lambda-t-list '0.001' \
--tta \
--tta-cfg-path ./configs/segm_release/_base_/tta_all_crop.py \
--save-data-name-list 'pred_seg_map' \
--job-name inference \
--partition gpu \
--num-gpus-per-node 1 \
--cpus-per-task 16 \
--array-parallelism 5
```
#### Visualization
Pre-compute and evulate time series data
```bash
python scripts/visualization/pre_compute_time_series.py \
--result_root ./results/segm_release \
--job-name eval \
--partition gpu \
--num-gpus-per-node 1 \
--cpus-per-task 24 \
--array-parallelism 8
```
Visualize labeled results
```bash
streamlit run scripts/visualization/labeled.py -- --result_root ./results/segm_release --model_name epoch_100_all
```
Visualize time series results
```bash
streamlit run scripts/visualization/time_series.py -- --result_root ./results/segm_release --model_name epoch_100_all
```
### todos
- [ ] Add commands for non-slurm users
- [ ] Add more details for data set structures
- [x] Update citation
### Acknowledgement
- [mabulnaga/automatic-placenta-segmentation](https://github.com/mabulnaga/automatic-placenta-segmentation)
- [voxelmorph/voxelmorph](https://github.com/voxelmorph/voxelmorph)
### License
This repo is licensed under the MIT License and the copyright belongs to all authors - see the [LICENSE](https://github.com/firstmover/cr-seg/blob/master/LICENSE) file for details.
### Citation
```
@inproceedings{liu2023consistency,
title={Consistency Regularization Improves Placenta Segmentation in Fetal EPI MRI Time Series},
author={Liu, Yingcheng and Karani, Neerav and Abulnaga, S Mazdak and Xu, Junshen and Grant, P Ellen and Abaci Turk, Esra and Golland, Polina},
booktitle={International Workshop on Preterm, Perinatal and Paediatric Image Analysis},
pages={77--87},
year={2023},
organization={Springer}
}
```
### Contact
Email: [email protected]