https://github.com/gagniuc/local-sequence-alignment-in-js
This JavaScript implementation detects the areas where two DNA/RNA/protein sequences are similar to each other. All symbols from UTF-8 are accepted by this algorithm.
https://github.com/gagniuc/local-sequence-alignment-in-js
alignment dna javascript js local native proteins rna sequence similarity similarity-matrix similarity-score
Last synced: 2 months ago
JSON representation
This JavaScript implementation detects the areas where two DNA/RNA/protein sequences are similar to each other. All symbols from UTF-8 are accepted by this algorithm.
- Host: GitHub
- URL: https://github.com/gagniuc/local-sequence-alignment-in-js
- Owner: Gagniuc
- License: mit
- Created: 2021-11-13T20:02:07.000Z (almost 4 years ago)
- Default Branch: main
- Last Pushed: 2022-03-21T10:06:50.000Z (over 3 years ago)
- Last Synced: 2025-01-15T07:31:57.275Z (9 months ago)
- Topics: alignment, dna, javascript, js, local, native, proteins, rna, sequence, similarity, similarity-matrix, similarity-score
- Language: HTML
- Homepage:
- Size: 296 KB
- Stars: 3
- Watchers: 1
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- Funding: .github/FUNDING.yml
- License: LICENSE.md
Awesome Lists containing this project
README
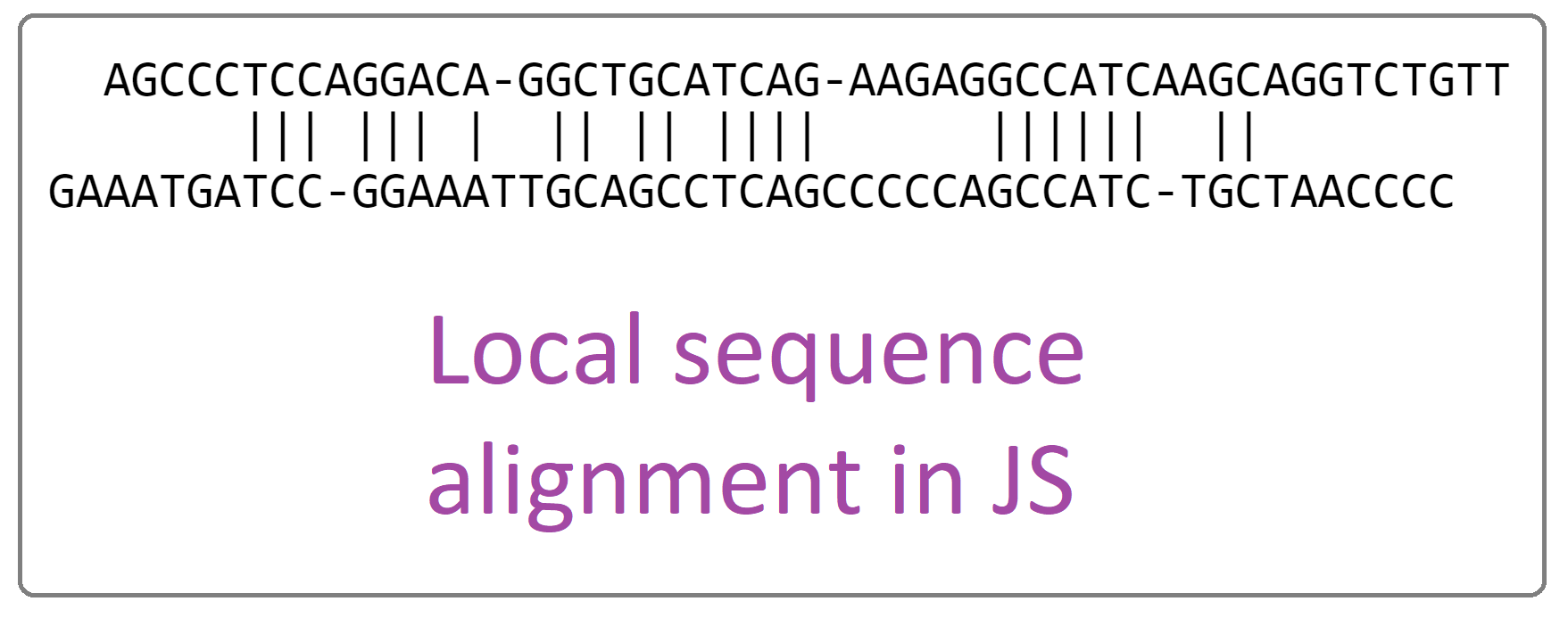
# Local sequence alignment in JS
This JavaScript implementation detects the areas where two DNA/RNA/protein sequences are similar to each other. In fact, this simple implementation makes an alignment between any two strings. All symbols from UTF-8 are accepted by this algorithm. Moreover, the implementation is the core of the application found here: https://github.com/Gagniuc/Jupiter-Bioinformatics-V2-normal
# Live demo
https://gagniuc.github.io/Local-sequence-alignment-in-JS/
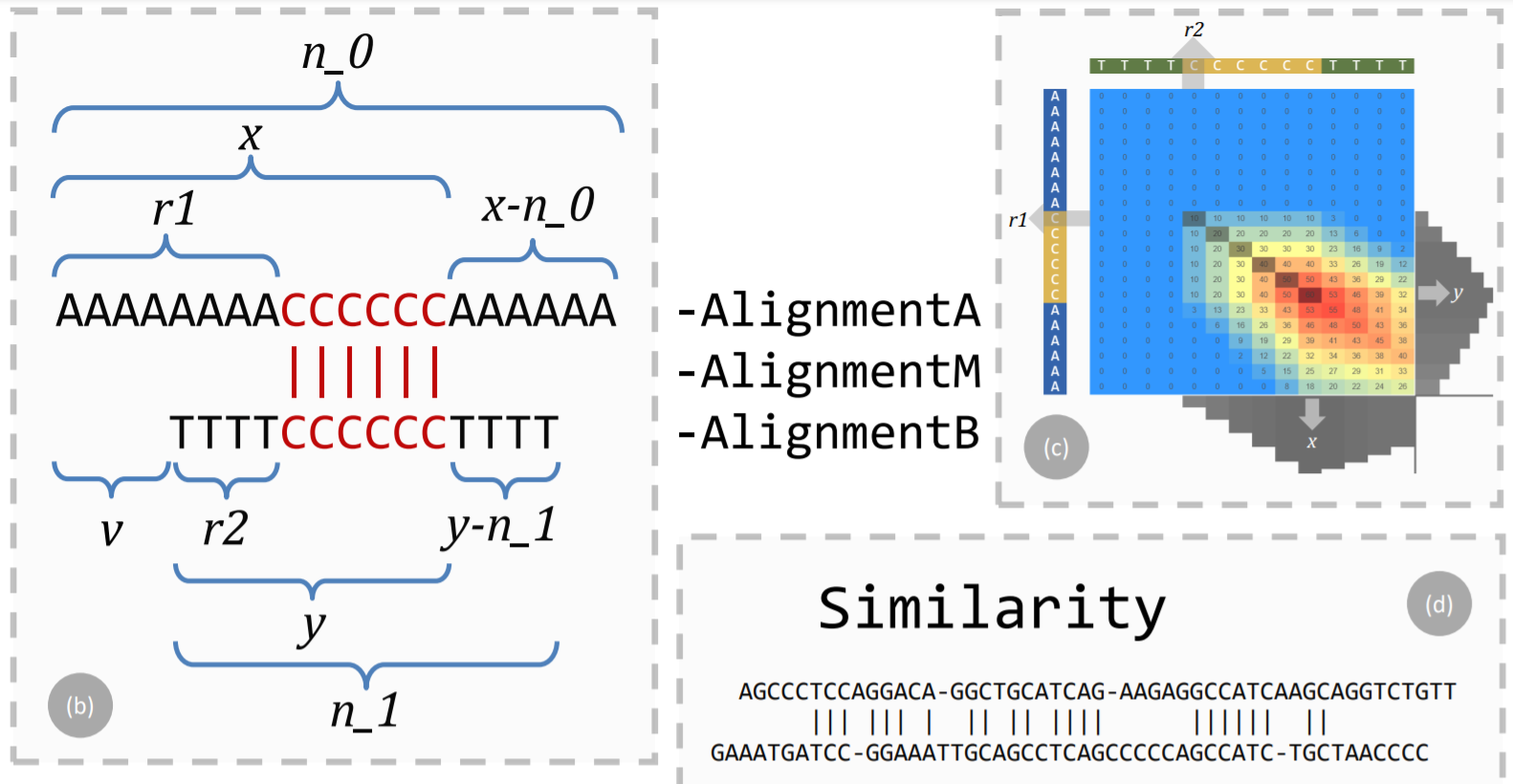
# Screenshot
# References
- Paul A. Gagniuc. Algorithms in Bioinformatics: Theory and Implementation. John Wiley & Sons, Hoboken, NJ, USA, 2021, ISBN: 9781119697961.