https://github.com/gbuesing/pca
Principal component analysis (PCA) in Ruby
https://github.com/gbuesing/pca
pca principal-component-analysis ruby rubyml
Last synced: 10 months ago
JSON representation
Principal component analysis (PCA) in Ruby
- Host: GitHub
- URL: https://github.com/gbuesing/pca
- Owner: gbuesing
- License: mit
- Created: 2015-03-31T23:17:26.000Z (almost 11 years ago)
- Default Branch: master
- Last Pushed: 2017-01-05T19:06:44.000Z (about 9 years ago)
- Last Synced: 2025-04-27T02:47:30.531Z (11 months ago)
- Topics: pca, principal-component-analysis, ruby, rubyml
- Language: Ruby
- Homepage:
- Size: 469 KB
- Stars: 26
- Watchers: 0
- Forks: 9
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- License: MIT-LICENSE
Awesome Lists containing this project
- data-science-with-ruby - pca
README
# Principal Component Analysis (PCA)
[Principal component analysis](http://setosa.io/ev/principal-component-analysis/) in Ruby. Uses [GSL](http://www.gnu.org/software/gsl/) for calculations.
PCA can be used to map data to a lower dimensional space while minimizing information loss.
It's useful for data visualization, where you're limited to 2-D and 3-D plots.
For example, here's a plot of the 4-D iris flower dataset mapped to 2-D via PCA:
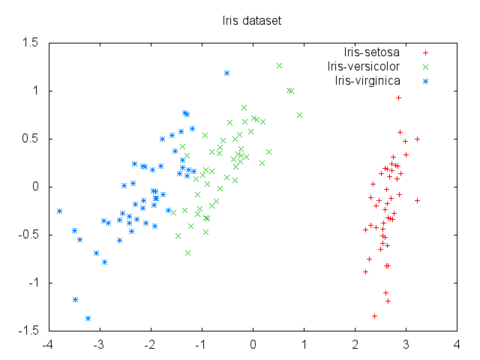
PCA is also used to compress the features of a dataset before feeding it into a machine learning algorithm,
potentially speeding up training time with a minimal loss of data detail.
## Install
**GSL must be installed first**. On OS X it can be installed via homebrew: ```brew install gsl```
gem install pca
## Example Usage
```ruby
require 'pca'
pca = PCA.new components: 1
data_2d = [
[2.5, 2.4], [0.5, 0.7], [2.2, 2.9], [1.9, 2.2], [3.1, 3.0],
[2.3, 2.7], [2.0, 1.6], [1.0, 1.1], [1.5, 1.6], [1.1, 0.9]
]
data_1d = pca.fit_transform data_2d
# Transforms 2d data into 1d:
# data_1d ~= [
# [-0.8], [1.8], [-1.0], [-0.3], [-1.7],
# [-0.9], [0.1], [1.1], [0.4], [1.2]
# ]
more_data_1d = pca.transform [ [3.1, 2.9] ]
# Transforms new data into previously fitted 1d space:
# more_data_1d ~= [ [-1.6] ]
reconstructed_2d = pca.inverse_transform data_1d
# Reconstructs original data (approximate, b/c data compression):
# reconstructed_2d ~= [
# [2.4, 2.5], [0.6, 0.6], [2.5, 2.6], [2.0, 2.1], [2.9, 3.1]
# [2.4, 2.6], [1.7, 1.8], [1.0, 1.1], [1.5, 1.6], [1.0, 1.0]
# ]
evr = pca.explained_variance_ratio
# Proportion of data variance explained by each component
# Here, the first component explains 99.85% of the data variance:
# evr ~= [0.99854]
```
See [examples](examples/) for more. Also, peruse the [source code](lib/pca.rb) (~ 100 loc.)
### Options
The following options can be passed in to ```PCA.new```:
option | default | description
------ | ------- | -----------
:components | nil | number of components to extract. If nil, will just rotate data onto first principal component
:scale_data | false | scales features before running PCA by dividing each feature by its standard deviation.
### Working with Returned GSL::Matrix
```PCA#transform```, ```#fit_transform```, ```#inverse_transform``` and ```#components``` return instances of ```GSL::Matrix```.
Some useful methods to work with these are the ```#each_row``` and ```#each_col``` iterators,
and the ```#row(i)``` and ```#col(i)``` accessors.
Or if you'd prefer to work with a standard Ruby ```Array```, you can just call ```#to_a``` and get an array of row arrays.
See [GSL::Matrix RDoc](http://blackwinter.github.io/rb-gsl/rdoc/matrix_rdoc.html) for more.
### Plotting Results With GNUPlot
Requires [GNUPlot](http://www.gnuplot.info/) and [gnuplot gem](https://github.com/rdp/ruby_gnuplot/tree/master).
```ruby
require 'pca'
require 'gnuplot'
pca = PCA.new components: 2
data_2d = pca.fit_transform data
Gnuplot.open do |gp|
Gnuplot::Plot.new(gp) do |plot|
plot.title "Transformed Data"
plot.terminal "png"
plot.output "out.png"
# Use #col accessor to get separate x and y arrays
# #col returns a GSL::Vector, so be sure to call #to_a before passing to DataSet
xy = [data_2d.col(0).to_a, data_2d.col(1).to_a]
plot.data << Gnuplot::DataSet.new(xy) do |ds|
ds.title = "Points"
end
end
end
```
## Sources and Inspirations
- [A tutorial on Principal Components Analysis](http://www.cs.otago.ac.nz/cosc453/student_tutorials/principal_components.pdf) (PDF) a great introduction to PCA
- [Principal Component Analyisis Explained Visually](http://setosa.io/ev/principal-component-analysis/)
- [scikit-learn PCA](http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html)
- [Lecture video](https://www.coursera.org/learn/machine-learning/lecture/ZYIPa/principal-component-analysis-algorithm) and [notes](https://share.coursera.org/wiki/index.php/ML:Dimensionality_Reduction) (requires Coursera login) from Andrew Ng's Machine Learning Coursera class
- [Implementing a Principal Component Analysis (PCA) in Python step by step](http://sebastianraschka.com/Articles/2014_pca_step_by_step.html)
- [Dimensionality Reduction: Principal Component Analysis in-depth](http://nbviewer.ipython.org/github/jakevdp/sklearn_pycon2015/blob/master/notebooks/04.1-Dimensionality-PCA.ipynb)