Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/ggseg/python-ggseg
Python module for ggseg-like visualizations
https://github.com/ggseg/python-ggseg
Last synced: 1 day ago
JSON representation
Python module for ggseg-like visualizations
- Host: GitHub
- URL: https://github.com/ggseg/python-ggseg
- Owner: ggseg
- License: mit
- Created: 2021-11-10T18:53:31.000Z (about 3 years ago)
- Default Branch: main
- Last Pushed: 2021-12-26T15:07:36.000Z (almost 3 years ago)
- Last Synced: 2024-11-09T17:45:35.317Z (4 days ago)
- Language: Python
- Homepage:
- Size: 1.49 MB
- Stars: 99
- Watchers: 2
- Forks: 16
- Open Issues: 3
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README

[](https://coveralls.io/github/ggseg/python-ggseg?branch=main)
[](https://github.com/ggseg/python-ggseg/blob/main/LICENSE)
[](https://pypi.org/project/ggseg/)
# python-ggseg
Python module for ggseg-like visualizations.
## Dependencies
Requires `matplotlib>=3.4` and `numpy>=1.21`.
## Install
```pip install ggseg```
## Usage
In order to work with `python-ggseg`, the data should be prepared as a
dictionary where each item is one region of a given atlas assigned with some
numeric value. The current version includes three atlases: the
[Desikan-Killiany (DK) atlas](https://pubmed.ncbi.nlm.nih.gov/16530430/)
, the [Johns Hopkins University (JHU) white-matter atlas](https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/Atlases) and the [FreeSurfer `aseg` atlas](https://surfer.nmr.mgh.harvard.edu/fswiki/FreeSurferVersion3).
### Cortical ROIs (Desikan-Killiany)
Cortical ROI data such as using the DK atlas may be
structured as follows:
> ```
> {'bankssts_left': 1.1,
> 'caudalanteriorcingulate_left': 1.0,
> 'caudalmiddlefrontal_left': 2.6,
> 'cuneus_left': 2.6,
> 'entorhinal_left': 0.6,
> ...}
Then be passed to the `ggseg.plot_dk` function:
```python
import ggseg
ggseg.plot_dk(data, cmap='Spectral', figsize=(15,15),
background='k', edgecolor='w', bordercolor='gray',
ylabel='Cortical thickness (mm)', title='Title of the figure')
```
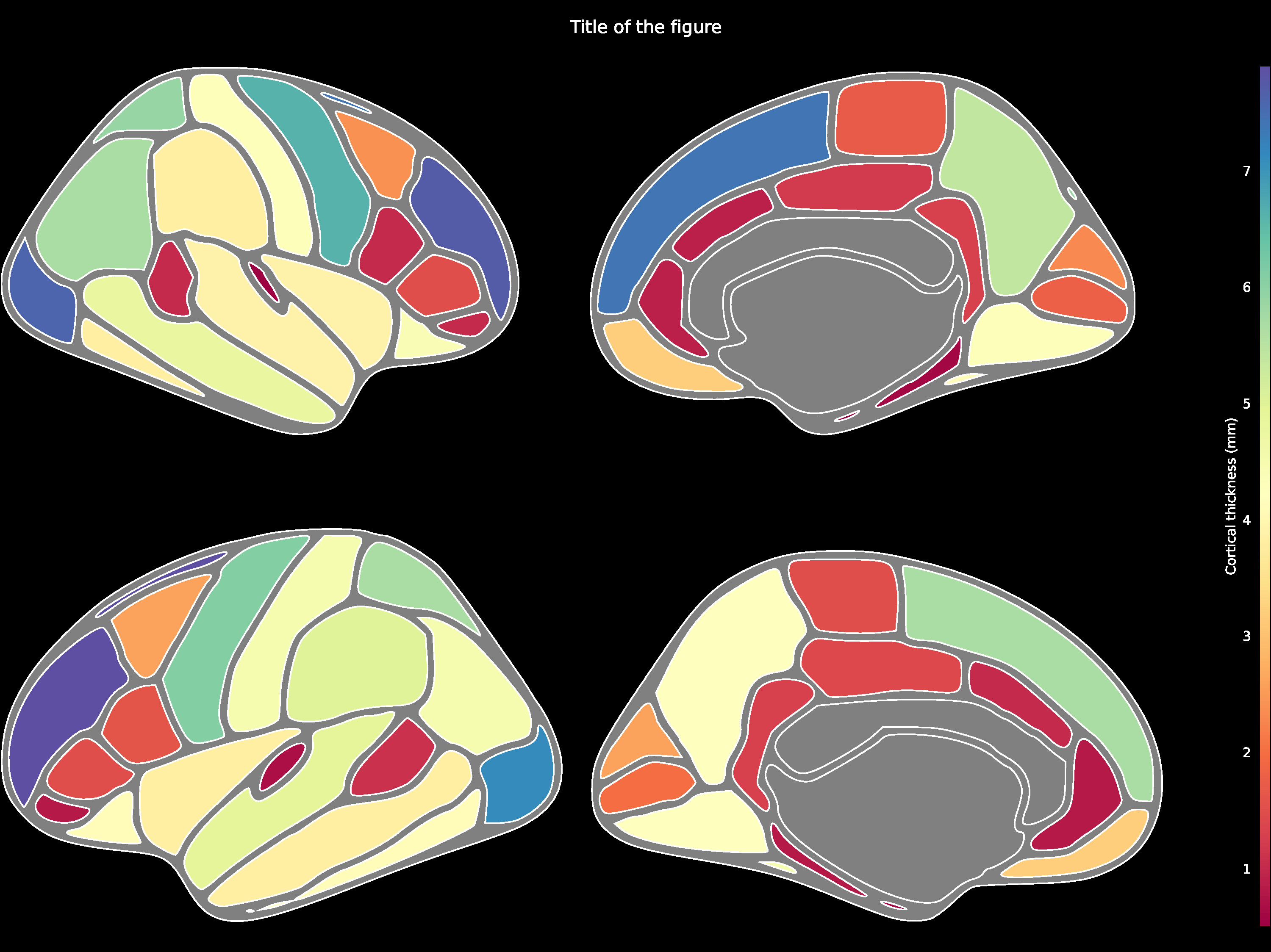
The comprehensive list of applicable regions can be found in this [folder](https://github.com/ggseg/python-ggseg/tree/main/ggseg/data/dk).
### Subcortical regions (FreeSurfer `aseg` atlas)
```python
data = {'Left-Lateral-Ventricle': 12289.6,
'Left-Thalamus': 8158.3,
'Left-Caudate': 3463.3,
'Left-Putamen': 4265.3,
'Left-Pallidum': 1620.9,
'3rd-Ventricle': 1635.6,
'4th-Ventricle': 1115.6,
...}
```
```python
ggseg.plot_aseg(data, cmap='Spectral',
background='k', edgecolor='w', bordercolor='gray',
ylabel='Volume (mm3)', title='Title of the figure')
```
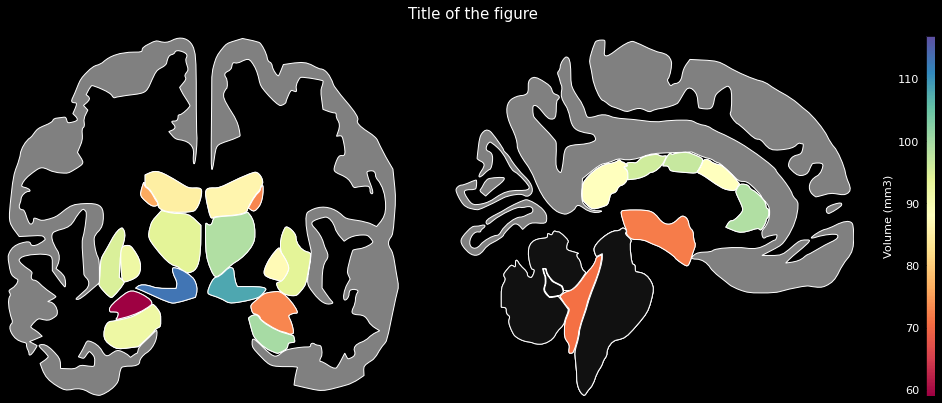
The comprehensive list of applicable regions can be found in this [folder](https://github.com/ggseg/python-ggseg/tree/main/ggseg/data/aseg).
### White-matter fiber tracts (Johns Hopkins University)
```python
data = {'Anterior thalamic radiation L': 0.3004812598228455,
'Anterior thalamic radiation R': 0.2909256815910339,
'Corticospinal tract L': 0.3517134189605713,
'Corticospinal tract R': 0.3606771230697632,
'Cingulum (cingulate gyrus) L': 0.3149917721748352,
'Cingulum (cingulate gyrus) R': 0.3126821517944336,
...}
```
```python
ggseg.plot_jhu(data, background='k', edgecolor='w', cmap='Spectral',
bordercolor='gray', ylabel='Mean Fractional Anisotropy',
title='Title of the figure')
```
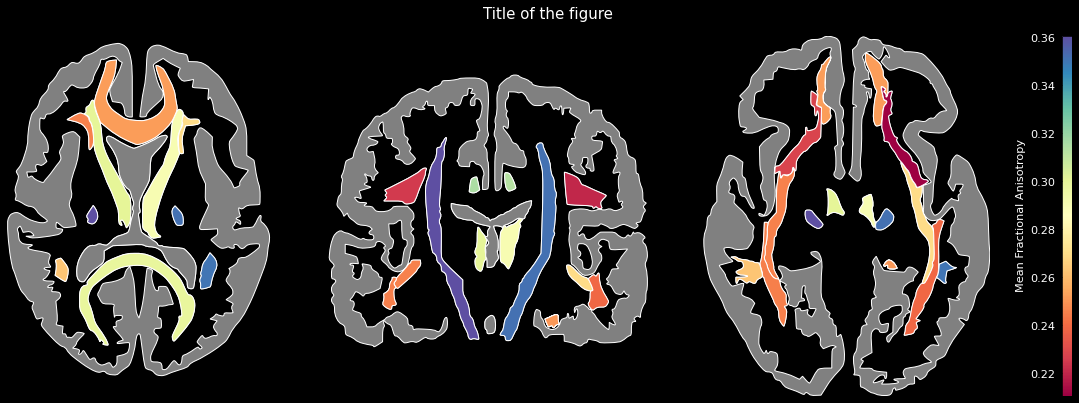
The comprehensive list of applicable regions can be found in this [folder](https://github.com/ggseg/python-ggseg/tree/main/ggseg/data/jhu).
## Tests
The current development version of `python-ggseg` has a coverage rate close to 100%.
The corresponding tests can be found in this [folder](https://github.com/ggseg/python-ggseg/tree/main/ggseg/tests).
## Examples
A Jupyter Notebook with examples can be found [there](https://github.com/ggseg/python-ggseg/blob/main/doc/ggseg.ipynb).