https://github.com/giovcandido/swarming
Swarming is a Python3 library that features both parallel and serial implementation of the Particle Swarm Optimization (PSO) algorithm.
https://github.com/giovcandido/swarming
distributed-computing library metaheuristic optimization optimizer parallel particle-swarm-optimization pso python3 swarm-optimization-algorithm
Last synced: 5 months ago
JSON representation
Swarming is a Python3 library that features both parallel and serial implementation of the Particle Swarm Optimization (PSO) algorithm.
- Host: GitHub
- URL: https://github.com/giovcandido/swarming
- Owner: giovcandido
- License: gpl-3.0
- Created: 2021-07-02T19:20:50.000Z (over 4 years ago)
- Default Branch: master
- Last Pushed: 2021-10-26T20:26:49.000Z (about 4 years ago)
- Last Synced: 2025-06-10T07:09:34.651Z (6 months ago)
- Topics: distributed-computing, library, metaheuristic, optimization, optimizer, parallel, particle-swarm-optimization, pso, python3, swarm-optimization-algorithm
- Language: Python
- Homepage:
- Size: 449 KB
- Stars: 7
- Watchers: 1
- Forks: 2
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Swarming - PSO and ParallelPSO Python3 library
[![PyPI][pypi-badge]][pypi-link]
[![PyPI - Downloads][install-badge]][install-link]
[![PyPI - Status][status-badge]][status-link]
[![PyPI - License][license-badge]][license-link]
Optimize as much as you want with this easy-to-use library.
## Contents
- [About](#about)
- [Usage](#usage)
- [Constructors](#constructors)
- [Working](#working)
- [Illustration](#illustration)
- [Parser](#parser)
- [Output](#output)
- [Examples](#examples)
- [Installation](#installation)
- [Acknowledgement](#acknowledgement)
- [Contribute](#contribute)
## About
Swarming is a Python3 library that features both parallel and serial
implementation of the Particle Swarm Optimization (PSO) algorithm.
It's made with Python3 and tested on Linux.
## Usage
In order to use Swarming for an optimization task, you need to choose
between PSO and ParallelPSO. It's important to note that the further one
works better for long calculations.
### Constructors
After making your choice, you're ready to go. Both PSO and ParallelPSO
constructors take 5 arguments. The arguments are:
- swarm_size - the number of particles in the swarm;
- dimension - the number of function variables;
- function - the function you wish to optimize;
- lower_bounds - the lower bounds of the function variables;
- upper_bounds - the upper bounds of the function variables.
### Working
Bear in mind that for this library optimizing a function means finding a
point of minimum function value. In the PSO algorithm, the particles
represent candidate solutions to the optimization problem.
The particles are randomly placed within the search space boundaries and move
in the direction that is more convenient for a certain number of iterations. By
the end of the task, it's expected that the swarm gets to an optimum position.
It is essential to discuss the algorithm metric for measuring "convenience".
The algorithm not only keeps track of the position of the particles, but of
the best particle position and of the best position of the whole swarm.
In short, the particle position, its best position and the best swarm position
are taken into consideration when calculating the "direction" a particular
particle should go to in the search space.
The best particle position and the best swarm position are both determined by
the function value in the particle position, that is the particle score. Since
the algorithm tries finding a minimum value, the lower the score, the better.
Having said all that, the function to be optimized must receive a single
argument, that is a point or position in the space. Additionally, it needs
to return one single value, that is the function value on the passed point.
### Illustration
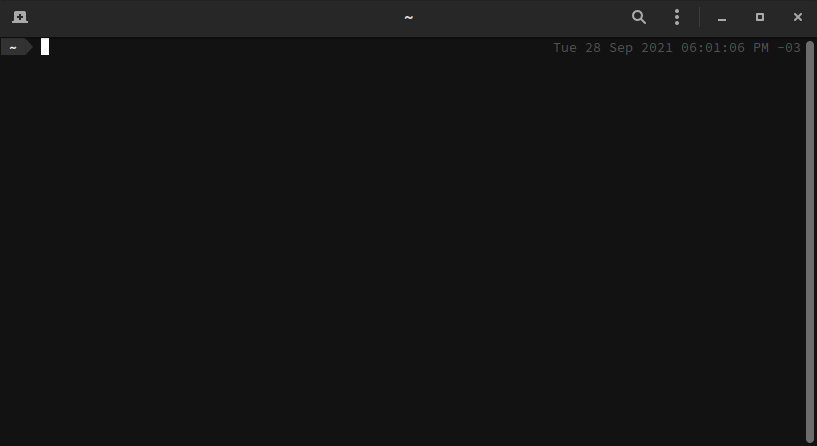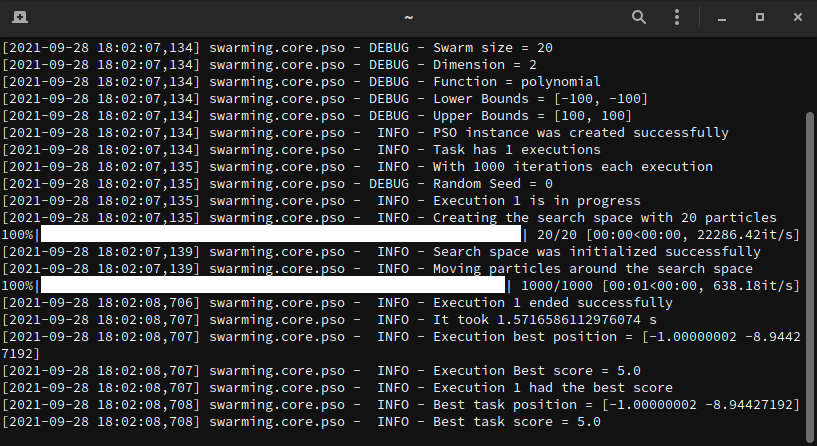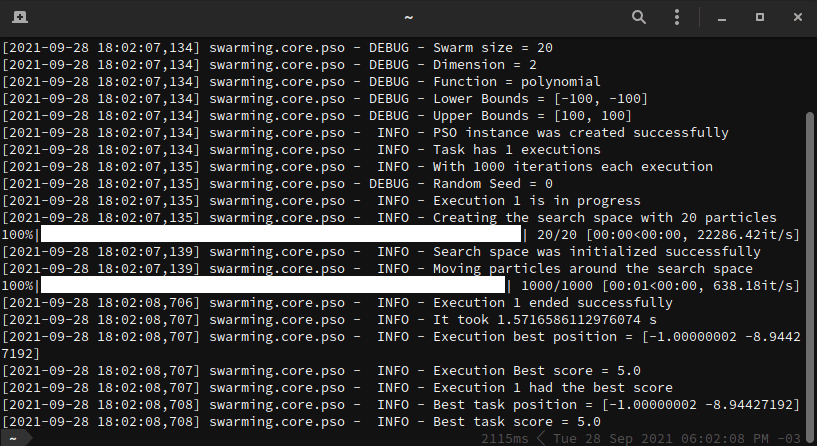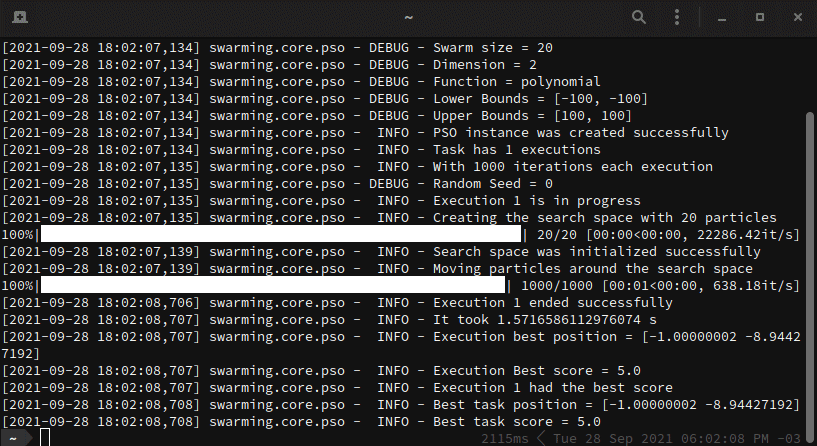
Below, we have a working example. In this example, we want to optimize a
polynomial function. For this task, let's consider 20 particles moving in a
restricted search space, that is bounded by [-100, 100] and [100, 100], for
a total of 1000 iterations.
```python
from swarming.core.pso import PSO
def polynomial(x):
return x[0] ** 2 + ((x[1] ** 2) / 16 - 5) ** 2 + 2 * x[0] + 6
swarm_size = 20
dimension = 2
lower_bounds = [-100, -100]
upper_bounds = [100, 100]
pso = PSO(swarm_size, dimension, polynomial, lower_bounds, upper_bounds)
pso.optimize(iterations=1000)
```
Notice that optimizing a polynomial function doesn't require long computations,
so we chose to use The PSO class instead of the ParallelPSO. But, in case you
want to test the above example with the ParallelPSO class, you just need to
replace the PSO with ParallelPSO in the code.
Making it easier for you, the example with ParallelPSO would be as follows:
```python
from swarming.core.pso import ParallelPSO
def polynomial(x):
return x[0] ** 2 + ((x[1] ** 2) / 16 - 5) ** 2 + 2 * x[0] + 6
swarm_size = 20
dimension = 2
lower_bounds = [-100, -100]
upper_bounds = [100, 100]
pso = ParallelPSO(swarm_size, dimension, polynomial, lower_bounds, upper_bounds)
pso.optimize(iterations=1000)
```
### Parser
Intending to facilitate the execution of a certain task, Swarming also provides
an argument parser. The parser is supposed to help you experiment with the same
function to be optimized. You can quickly try different settings.
The parser has four predefined arguments:
- (P) if it should run in parallel;
- (S) the number particles in the swarm;
- (I) the number of iterations in the task;
- (E) the number of executions of the task.
Using the parser, you could execute a certain task by running:
```bash
python3 example.py --parallel --swarm-size S --iterations I --executions E
```
You can also run:
```bash
python3 example.py -p -s S -i I -e E
```
Note that you should __replace__ example.py with the desired script and the
uppercase letters with the values you wish.
Moreover, it's also important to note that 'parallel' and 'executions' are
optional arguments. If you don't include them when executing a script, the
default values are considered. For parallel, the default is 'False', which
means no parallel execution. As for executions, the default is one execution.
If you are wondering how an optimization script would be with the parser,
here goes an example for you:
```python
from swarming.utils.argument_parser import parse_arguments
from swarming.core.pso import PSO, ParallelPSO
def polynomial(x):
return x[0] ** 2 + ((x[1] ** 2) / 16 - 5) ** 2 + 2 * x[0] + 6
args = parse_arguments()
parallel = args.parallel
swarm_size = args.swarm_size
iterations = args.iterations
executions = args.executions
dimension = 2
lower_bounds = [-100, -100]
upper_bounds = [100, 100]
if not parallel:
PSOClass = PSO
else:
PSOClass = ParallelPSO
pso = PSOClass(swarm_size, dimension, polynomial, lower_bounds, upper_bounds)
pso.optimize(iterations=iterations, executions=executions)
```
### Output
Every execution generates one unique __.log__ file that describes step-by-step what happens in the optimization task.
Additionally, one unique __.npy__ is also saved. This file contains the best task position, that is the solution to the optimization problem.
In order to load the solution, all you have to do is:
```python3
import numpy as np
sol = np.load('file.npy')
```
## Examples
In the examples directory, there are currently three scripts:
- polynomial.py;
- exponential.py;
- neural_net.py.
You can pick any script to test. Moreover, you can change them as you want.
In order to test Swarming for the first time, keep the scripts unchanged
and execute them.
The first two scripts don't not use the parser, while the latter does.
So, the first script can be executed as follows:
```bash
python3 polynomial.py
```
The third script can be executed this way:
```bash
python3 neural_net.py --swarm-size 10 --iterations 100
```
## Installation
There are two ways you can install Swarming: you can install it from source or
you can get it using the pip3 command.
If you want to get it from source, download the latest release on GitHub or
clone the repository. Then, extract the source code and run:
```bash
pip3 install -e .
```
If you want to install Swarming with pip, you just need to run:
```bash
pip3 install swarming
```
You can also run:
```bash
sudo pip3 install swarming
```
## Acknowledgement
The idea of Particle Swarm Optimization can be found in
[Kennedy's paper](https://ieeexplore.ieee.org/document/488968).
It's worth mentioning that my colleague
[gugarosa](https://github.com/gugarosa) made sure I was on the right path.
## Contribute
Feel free to reach out and contribute. We can add more features to Swarming.
Furthermore, if you have any problem or suggestion, open an issue.
If you want to talk to us, send a message to giovcandido@outlook.com.
[pypi-badge]: https://img.shields.io/pypi/v/swarming.svg
[pypi-link]: https://pypi.org/project/swarming
[install-badge]: https://img.shields.io/pypi/dm/swarming?label=pypi%20installs
[install-link]: https://pypistats.org/packages/swarming
[license-badge]: https://img.shields.io/pypi/l/swarming.svg
[license-link]: https://pypi.python.org/pypi/swarming/
[status-badge]: https://img.shields.io/pypi/status/swarming.svg
[status-link]: https://pypi.python.org/pypi/swarming/