Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/grburgess/gbm_kitty
Database, reduce, and analyze GBM data without having to know anything. Curiosity killed the catalog.
https://github.com/grburgess/gbm_kitty
3ml catalogue data-analysis fermi-science grbs pipelines
Last synced: 13 days ago
JSON representation
Database, reduce, and analyze GBM data without having to know anything. Curiosity killed the catalog.
- Host: GitHub
- URL: https://github.com/grburgess/gbm_kitty
- Owner: grburgess
- License: gpl-3.0
- Created: 2021-01-08T07:00:59.000Z (about 4 years ago)
- Default Branch: main
- Last Pushed: 2021-01-13T14:01:55.000Z (about 4 years ago)
- Last Synced: 2024-11-23T15:49:20.494Z (2 months ago)
- Topics: 3ml, catalogue, data-analysis, fermi-science, grbs, pipelines
- Language: Python
- Homepage:
- Size: 14.8 MB
- Stars: 2
- Watchers: 1
- Forks: 2
- Open Issues: 1
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README

GBM Kitty
Database, reduce, and analyze GBM data without having to know anything. Curiosity killed the catalog.
## What is this?
* Creates a MongoDB database of GRBs observed by GBM.
* Heuristic algorithms are applied to search for the background regions in the time series of GBM light curves.
* Analysis notebooks can be generated on the fly for both time-instegrated and time-resolved spectral fitting.
Of course, this analysis is highly opinionated.
## What this is not
Animal cruelty.
## What can you do?
Assuming you have built a local database (tis possible, see below), just type:
```bash
$> get_grb_analysis --grb GRBYYMMDDxxx
```

magic happens, and then you can look at your locally built GRB analysis notebook.
If you want to do more, go ahead and fit the spectra:
```bash
$> get_grb_analysis --grb GRBYYMMDDxxx --run-fit
```

And your automatic (but mutable) analysis is ready:
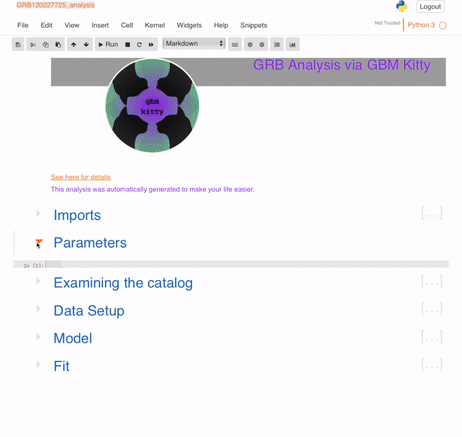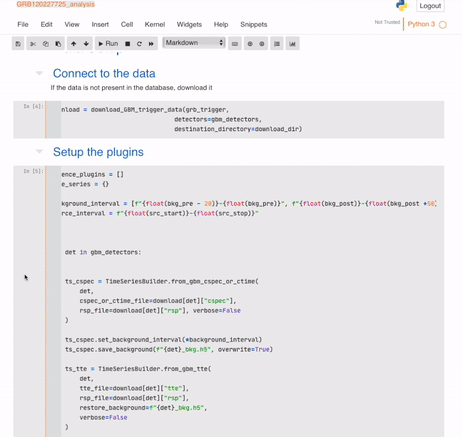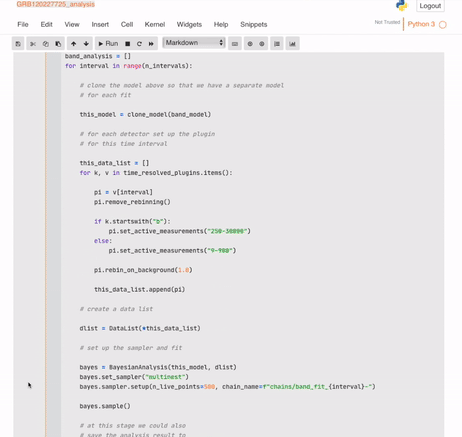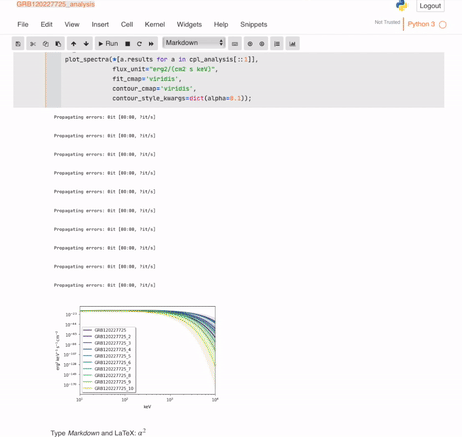
## Building the database
The concept behind this is to query the Fermi GBM database for basic trigger info, use this in combination tools such as [gbmgeometry](https://gbmgeometry.readthedocs.io/en/latest/) to figure out which detectors produce the best data for each GRB, and then figure out preliminary selections / parameters / setups for subsequent analysis.
```bash
$> build_catalog --n_grbs 100 --port 8989
```
This process starts with launching [luigi](https://luigi.readthedocs.io/en/stable/) which mangages the pipline:
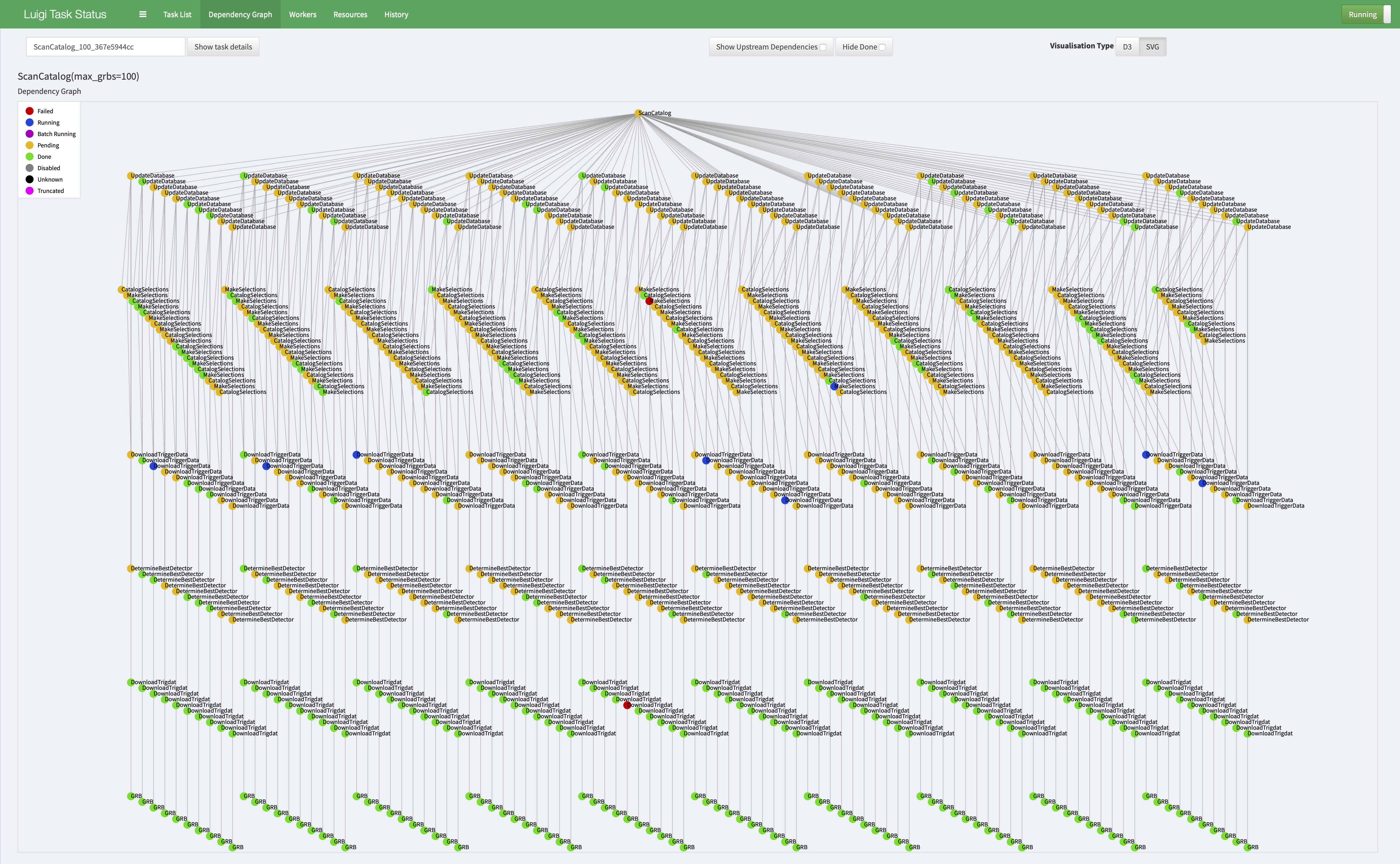
All the of the metadata about the process is stored in a [mondoDB](https://www.mongodb.com) database which can be referenced later when building analyses.