https://github.com/greenelab/continuous_analysis_brainarray
Continuous Analysis Example - Performing Differential Expression Analysis with Custom Chip Description Files (CustomCDF)
https://github.com/greenelab/continuous_analysis_brainarray
analysis continuous-integration example methodology workflow
Last synced: 11 months ago
JSON representation
Continuous Analysis Example - Performing Differential Expression Analysis with Custom Chip Description Files (CustomCDF)
- Host: GitHub
- URL: https://github.com/greenelab/continuous_analysis_brainarray
- Owner: greenelab
- License: bsd-3-clause
- Created: 2016-08-08T15:26:37.000Z (over 9 years ago)
- Default Branch: master
- Last Pushed: 2016-12-21T19:49:49.000Z (about 9 years ago)
- Last Synced: 2025-01-13T00:41:36.535Z (about 1 year ago)
- Topics: analysis, continuous-integration, example, methodology, workflow
- Language: R
- Homepage: http://dx.doi.org/10.1101/056473
- Size: 1.43 MB
- Stars: 0
- Watchers: 5
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Continuous Analysis BrainArray CustomCDF Example
This is a sample repository showing a [Continuous Analysis Workflow](https://github.com/greenelab/continuous_analysis) for RNA-Seq analysis. A description of continuous analysis is available as a [pre-print](http://dx.doi.org/10.1101/056473).
In this example we evaluate the effect of different custom chip description files (CustomCDF). To evaluate the impact of differing CDF versions, we downloaded a recently published public gene expression dataset. This experiment examined differential expression between normal HeLa cells and HeLa cells with TIA1 and TIAR knocked down ([GEO Series Ascension number GSE47664](http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47664)). We performed a parallel analysis using each of the three versions that we found installed on machines that we could access (18, 19, and 20). Each version identifies a different number of significantly altered genes, demonstrating the challenge of reproducible analysis.
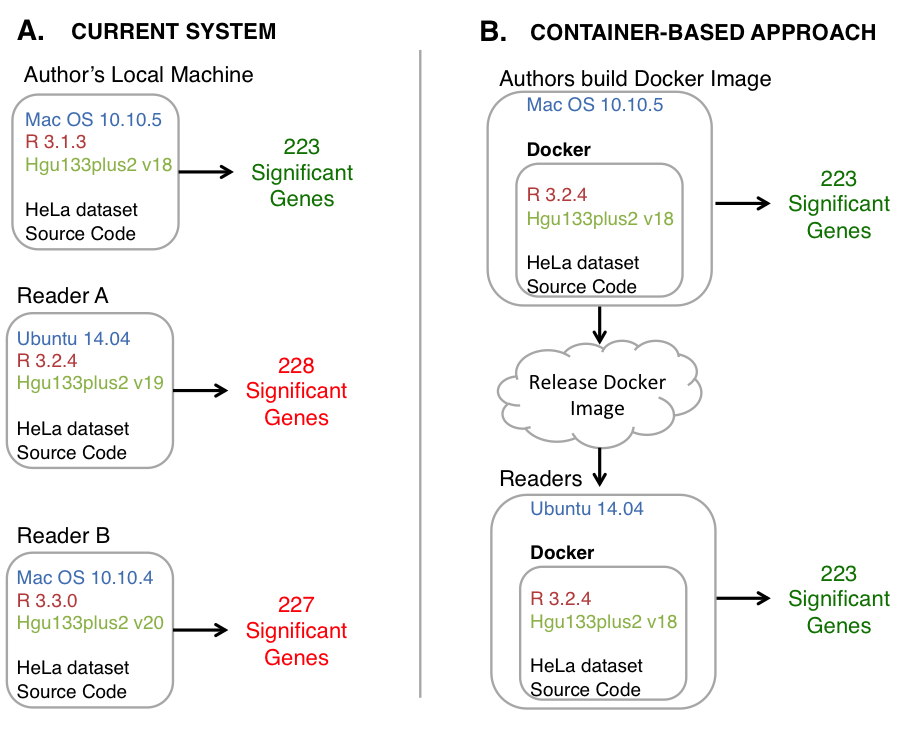
*Figure:* Current state of research computing vs. container-based approaches. A.) The status quo requires a reader or reviewer to find and install specific versions of dependencies. These dependencies can become difficult to find and may become incompatible with newer versions of other software packages. Different versions of packages identify different numbers of significantly differentially expressed genes from the same source code and data. B.) Containers define a computing environment that captures dependencies. In container-based systems, the results are the same regardless of the host system.
### Results:
The truncated output shows the first 100 genes for comparison.
[V19 -> V20]
Change: (https://github.com/greenelab/continuous_analysis_brainarray/commit/c969c281c9a55b8418a2115e74c1ff010bd19d86)
Result:(https://github.com/greenelab/continuous_analysis_brainarray/commit/443e8123ca9baa0b72d03c23dc07933ba1a3b5de)
[V18 -> V19]
Change: (https://github.com/greenelab/continuous_analysis_brainarray/commit/fc782b9dcc16a60f828cf94e597825b3fcec1513)
Result:(https://github.com/greenelab/continuous_analysis_brainarray/commit/55a63b83b1ee53a89c61a2d7c831f6ad74297620)
## Feedback
Please feel free to email me - (brettbe) at med.upenn.edu with any feedback or raise a github issue with any comments or questions.
## Acknowledgements
This work is supported by the Gordon and Betty Moore Foundation's Data-Driven Discovery Initiative through Grant GBMF4552 to C.S.G. as well as the Commonwealth Universal Research Enhancement (CURE) Program grant from the Pennsylvania Department of Health.