Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/grimmer0125/embedded-pydicom-react-viewer
Medical DICOM file P10 Viewer/Chrome Extension + Python Code In Browser (-Pyodide-> WebAssembly) + Pydicom parser + TypeScript React App (CRA). Use d4c-queue npm lib.
https://github.com/grimmer0125/embedded-pydicom-react-viewer
canvas dicom numpy nvm poetry pyenv pyodide python react typescript visualization webassembly
Last synced: 3 months ago
JSON representation
Medical DICOM file P10 Viewer/Chrome Extension + Python Code In Browser (-Pyodide-> WebAssembly) + Pydicom parser + TypeScript React App (CRA). Use d4c-queue npm lib.
- Host: GitHub
- URL: https://github.com/grimmer0125/embedded-pydicom-react-viewer
- Owner: grimmer0125
- License: other
- Created: 2021-04-03T17:15:57.000Z (almost 4 years ago)
- Default Branch: master
- Last Pushed: 2021-11-15T14:06:39.000Z (about 3 years ago)
- Last Synced: 2024-04-28T04:58:15.086Z (9 months ago)
- Topics: canvas, dicom, numpy, nvm, poetry, pyenv, pyodide, python, react, typescript, visualization, webassembly
- Language: Python
- Homepage: https://chrome.google.com/webstore/detail/dicom-image-viewer/ehppmcooahfnlfhhcflpkcjmonkoindc
- Size: 3.33 MB
- Stars: 21
- Watchers: 3
- Forks: 2
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE.md
Awesome Lists containing this project
README
# Embedded Pydicom React Viewer
This experimental project demonstrates
1. How to use Python in browser, working with ReactApp.
2. Use Python to parse DICOM files and pass data to JS, then draw it on Canvas.
Tested on macOS Big Sur (intel/M1), Chrome 89.
Its usage is simple. Just drag a DICOM file into the panel to view.
Download DICOM files from [DICOM sample file sites](#dicom-sample-file-sites)
## Features
- view DICOM files & show some basic info.
- window center & width mode
- multiple frame
- coronal & sagittal views & judge if current is AxialView or not
- scale (resize to viewer size)
- ...
more complete list is on https://github.com/grimmer0125/dicom-web-viewer/wiki and
(0028,3000) Modality LUT Sequence present DICOM & PALETTE COLOR are already supported in this project.
The Chrome extension is published, click [there](https://chrome.google.com/webstore/detail/dicom-image-viewer/ehppmcooahfnlfhhcflpkcjmonkoindc) to install.
## Motivation
Besides it is an interesting thing to use Python in browser, using Python DICOM parser has some advantages.
1. Although my another Chrome extension/Web project, https://github.com/grimmer0125/dicom-web-viewer uses 3-party JavaScript DICOM parser library but it seems not manintained. The other JavaScript/TypeScript DICOM parser library might be too heavy to use.
2. Scientists usually use Python DICOM parser library, and using the same language/library is a good thing.
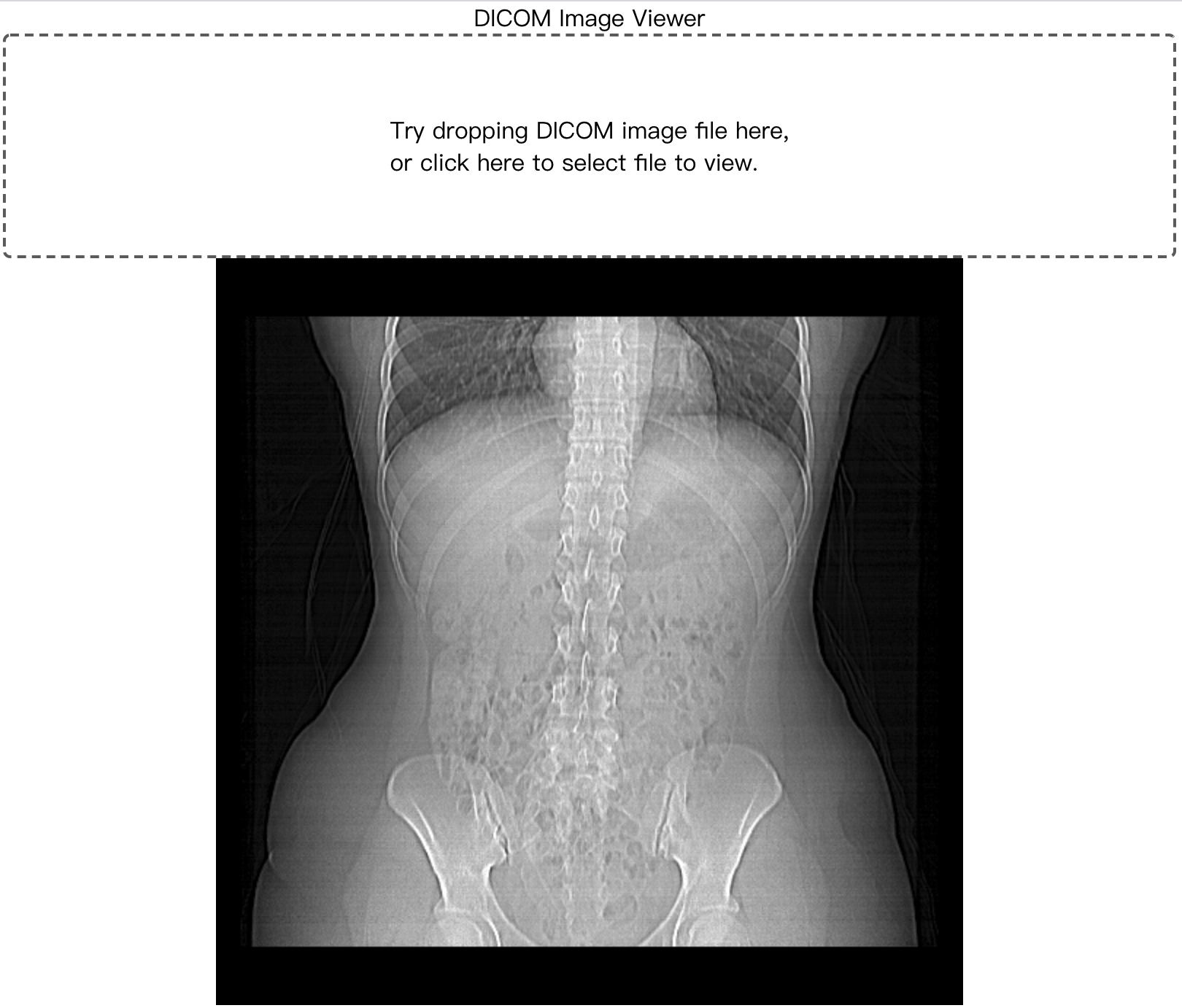
## Screenshot
OT-MONO2-8-hip.dcm from https://barre.dev/medical/samples/
## Python 3.9.5 Browser runtime - Pyodide 0.18.0
ref:
1. https://github.com/pyodide/pyodide
2. https://pyodide.org/en/latest/development/new-packages.html
### Other GitHub repos using Pyodide + Pydicom
1. [https://github.com/Fincap/onko-pyodide](https://github.com/Fincap/onko-pyodide), draw canvas in Pyodide runtime
2. [https://github.com/pymedphys/pymedphys](https://github.com/pymedphys/pymedphys), mainly for DICOM-RT
## Development
Please use VS Code and bulit-in TypeScript/Python formatter setting. Please install Python autopep8 out of thie project environment and mare sure the VS Code setting. Also, you can enable "format on save".
### Setup Pyodide
The current code uses local built Pyodide 0.18.0 version to speed up loading instead of CDN, just download it once. The zip file is https://github.com/grimmer0125/embedded-pydicom-react-viewer/releases/download/untagged-77e55ea5908aaad294e1/pyodide.zip and you can just execute
`$ sh download_pyodide.sh`
in terminal which will download+unzip+move to `public/pyodide`. These Pyodide fiels were download from `https://cdn.jsdelivr.net/`, not built from scratch.
Or you can comment these
```
await loadPyodide({ indexURL : "pyodide/" });
await micropip.install('pyodide/pydicom-2.2.1-py3-none-any.whl')
```
and replace by below to fetch from CDN
```
await loadPyodide({ indexURL : "https://cdn.jsdelivr.net/pyodide/dev/full/" });
await micropip.install('pydicom')
```
### Install Python, Node.js and their dependencies for intel and Mac M1 (arm) machines
https://github.com/nvm-sh/nvm January 2021: there are no pre-compiled NodeJS binaries for versions prior to 15.x for Apple's new M1 chip (arm64 architecture). v14.16 supports M1 but need compilation (auto done by nvm). p.s. nvm seems to still build 15.14.0
Make sure you have Node.js (v15.14.0+), Python (3.9.2+) and [Poetry](https://python-poetry.org/) installed first. (Optional) [pyenv](https://github.com/pyenv/pyenv) is recommended to switch different Python and it will automatically switch to 3.9.2 since .python-version is created.
Python is to help Python static code analysing and code completion and host built-react code. For example, you install pydicom from wheel url and use it in runtime but you can install pydicom (`poetry add pydicom -dev`) to help the auto type completion in VSCode.
Then
1. `git submodule update --init --recursive`
2. `npm install --global yarn`
3. `yarn install`
4. (optional) `poetry shell`
5. (optional) `poetry install`
### Start coding
Just `yarn start` (it may take a whlie for the 1st time since WebAssembly decoder is added)
## Production - Use Python FastAPI to host built React app
1. `yarn build` to build reactapp
2. To launch FastAPI,
either
```
$ poetry shell
$ uvicorn main:app
```
or
```
$ poetry run uvicorn main:app
```
Using `uvicorn main:app --reload` is for development but we already have create react app built-in development live server.
## Docker images
Building from latest code is not working now and wait for fix. `yarn build` happens out of memory error when building, and it probably is due to WebAssembly decoder.
### Build a docker image to run (either on amd64 or arm64)
1. `docker build --progress=plain -t pyodide-react-dicom-viewer .`
2. `docker run -p 8000:8000 -d pyodide-react-dicom-viewer`
3. open http://localhost:8000/ and drag a DICOM file to view.
### Build a universal docker image (supporting amd64/arm64)
Cross compliation for intel/m1 takes much more time than normal `docker build`. Building + Pushing to docker hub takes 20~30min. Several times.
1. `docker buildx create --use --name m1_builder`
2. `docker buildx use m1_builder`
3. `docker buildx inspect --bootstrap`
4. `docker buildx build --platform linux/amd64,linux/arm64 --push -t grimmer0125/pyodide-react-dicom-viewer:0.5 .`
### Use remote docker image to run
Image: https://hub.docker.com/repository/docker/grimmer0125/pyodide-react-dicom-viewer
1. `docker run -p 8000:8000 grimmer0125/pyodide-react-dicom-viewer:0.5`
2. open http://localhost:8000/ and drag a DICOM file to view.
## DICOM sample file sites
- https://barre.dev/medical/samples/ contains jpeg 57, 70 (MR-MONO2-12-shoulder, CT-MONO2-16-chest)
- pydicom lib
- https://github.com/pydicom/pydicom-data/tree/master/data_store/data JPGLosslessP14SV1_1s_1f_8b.dcm 1.2.840.10008.1.2.4.70 JPEG Lossless
- https://github.com/pydicom/pydicom/tree/master/pydicom/data/test_files jpeg 51
- http://www.rubomedical.com/dicom_files/, some (multi-frame) `DICOM jpeg 1.2.840.10008.1.2.4.50`
- https://medistim.com/dicom/
- http://medistim.com/wp-content/uploads/2016/07/ttfm.dcm 1.2.840.10008.1.2.4.70
- http://medistim.com/wp-content/uploads/2016/07/bmode.dcm ultra sound, 70, multi frame
- [gdcm](http://gdcm.sourceforge.net/wiki/index.php/Main_Page) data `git clone git://git.code.sf.net/p/gdcm/gdcmdata`
- ~~Daikon lib https://github.com/rii-mango/Daikon/tree/master/tests/data (these DICOM files miss transfersynax~~
### Tested sample files
Most of them are archived on https://github.com/grimmer0125/embedded-pydicom-react-viewer/releases/download/v0.2/dicom_samples.zip. ~~All jpeg compressed DICOM files need a extra JPEG decoder (except 50 baseline) to render on browser and currently it is parsed but not visible on browser. [Daikon][https://github.com/rii-mango/daikon] has done this, and https://github.com/cornerstonejs/dicomParser seems too.~~ The project already borrow the decoder from Dakon.
https://barre.dev/medical/samples/:
- CT-MONO2-16-ort: 1.2.840.10008.1.2, MONOCHROME2
- CR-MONO1-10-chest: 1.2.840.10008.1.2 (raw, need specified transfersyntax), MONOCHROME1
- US-RGB-8-esopecho: 1.2.840.10008.1.2.1, RGB, planar:0
- US-RGB-8-epicard: 1.2.840.10008.1.2.2, RRB, planar = 1
- JPEG57-MR-MONO2-12-shoulder: 1.2.840.10008.1.2.4.57 MONOCHROME2
- US-PAL-8-10x-echo: 1.2.840.10008.1.2.5 (RLE Lossless), PALETTE COLOR, multi-frame
https://github.com/pydicom/pydicom-data/tree/master/data_store/data
- color3d_jpeg_baseline: 1.2.840.10008.1.2.4.50, YBR_FULL_422 (not handled YBR part, so final contrast may be wrong), multi-frame
- JPGLosslessP14SV1_1s_1f_8b: 1.2.840.10008.1.2.4.70, MONOCHROME2
https://github.com/pydicom/pydicom/tree/master/pydicom/data/test_files
- JPEG-lossy: 1.2.840.10008.1.2.4.51, MONOCHROME2
- contrast of saved jpeg is not obvious
- JPEG2000: 1.2.840.10008.1.2.4.91, MONOCHROME2
- contrast of saved jpeg is not obvious
GDCM data, use `git://git.code.sf.net/p/gdcm/gdcmdata` to download
- D_CLUNIE_CT1_JLSL: 1.2.840.10008.1.2.4.80, MONOCHROME2 (saved jpeg-ls is not viewable on https://products.groupdocs.app/viewer/jpg, not sure it is normal or not)
- D_CLUNIE_CT1_JLSN: 1.2.840.10008.1.2.4.81, MONOCHROME2 (saved jpeg-ls is not viewable on https://products.groupdocs.app/viewer/jpg, not sure it is normal or not)
- DX_J2K_0Padding: 1.2.840.10008.1.2.4.90, MONOCHROME2
https://www.dclunie.com/images/compressed/index.html
- image_dfl: 1.2.840.10008.1.2.1.99, MONOCHROME2
## DICOM medical files - not handle cases
pydicom suported transfer syntax: https://pydicom.github.io/pydicom/dev/old/image_data_handlers.html
Below non handled items are done in another project https://github.com/grimmer0125/dicom-web-viewer (canvas operation is borrowed from this)
- DICOM FILE
- Main Transfer Syntax:
- [done] 51, 57, 70 JPEG DICOM.
- 1.2.840.10008.1.2.5 RLE Lossless (US-PAL-8-10x-echo.dcm is ok but not sure others)
- [done] 1.2.840.10008.1.2.4.80 JPEG LS Lossless
- [done] 1.2.840.10008.1.2.4.81 JPEG LS Lossy
- 1.2.840.10008.1.2.4.90 JPEG2000 Lossless
- [done] 1.2.840.10008.1.2.4.91 JPEG2000
- [done] 1.2.840.10008.1.2.1.99 Deflated Explicit VR Little Endian
- [done] Photometric: MONOCHROME1, inverted color
- [done] Photometric: RGB with planar 0, 1
- [done] Photometric: PALETTE
- [done] 1.2.840.10008.1.2.1 Explicit VR, Little Endian
- [done] 1.2.840.10008.1.2.2 Explicit VR, Big Endian
Transfer Syntax for videos (1.2.840.10008.1.2.4.100 / 1.2.840.10008.1.2.4.102 / 1.2.840.10008.1.2.4.103) and some other not often seen syntax will not be handled.
ref https://www.dicomlibrary.com/dicom/transfer-syntax/
## Issues
### performance
[Solved][performance] Using Python numpy in browser is slow in some cases (see below **Speed test**), it takes `3~4s` for 1 512\*512 array operation. Using pure JavaScript takes less than 0.5s. Ref: https://github.com/pyodide/pyodide/issues/112 (the author said WebAssembly may takes `3~5x` slow). The solution might be
1. (can rollback to git commit: `219299f9adec489134206faf0cfab79d8345a7df`), using pydicom to parse DICOM files, sending pixel data to JS, then use JS to flatten 2D grey data to 1D RGBA canvas image data.~~
2. [Use this way, solved] Or is there any quick way in numpy for flattening a 2D grey array to 1D RGBA array with normalization? Such as https://stackoverflow.com/questions/59219210/extend-a-greyscale-image-to-fit-a-rgb-image? Also image2D.min()/max() is fast. Need more study/profiling.
**Speed test (using above sample file to test, file: `OT-MONO2-8-hip.dcm` on https://barre.dev/medical/samples/)**:
1. numpy array + manual iteration calculation in local python ~= numpy array + numpy array operation ~= JS ArrayBuffer/int8ClampedArray + manual iteration calculation (very fast) >>
2. Python list + manual iteration calculation > (5s)
3. numpy array + manual iteration calculation in pyodide. (7s)
p.s.
1. I did not record JS accurate time cost but it is fast.
2. Local Python is much faster than Pyodide Python in browser.
## todo list
Besides adding back above medical file cases/features, there are some optional things we can do
1. [Done] Host these on your server. Check https://pyodide.org/en/0.17.0a2/usage/serving-pyodide-packages.html & https://pyodide.org/en/0.17.0a2/usage/loading-packages.html#
1. pyodide.wasm (WebAssembly, 10MB), pyodide.asm.js (3.8MB), and pyodide.asm.data(5MB) files
2. pyodide packages. e.g. numpy.js (159KB) and numpy.data (7.3MB <-used by WebAssembly). (By contrast, a numpy wheel package is about 16MB)
3. non pyodide built-in pure python packages (which needs to be a wheel package and we use `pyodide micropip` to install them from PyPI). e.g. pydicom-2.1.2-py3-none-any.whl (1.9MB)
2. Move python code to a browser webworker, https://pyodide.org/en/stable/usage/webworker.html.
3. [Done] Dockerization
4. [Done] Bundle some testing DICOM files
5. [Done] Introduction to medical files and pyodide
6. Make a Python package
7. 3D visualization
8. **Help to improve Pyodide
9. Refactor
10. Add tests
11. Fix [DICOM medical files - Not handle cases](#dicom-medical-files---not-handle-cases)
## Misc
This project is using my another TypeScript npm library, [d4c-queue](https://www.npmjs.com/package/d4c-queue), and code is on https://github.com/grimmer0125/d4c-queue/. You can take a look.