https://github.com/hicsail/constellation-js
Library for combinatorially specifying, constraining, and exploring genetic design spaces.
https://github.com/hicsail/constellation-js
combinatorial-search computational-biology constraint-programming genetic-design graph graph-algorithms javascript programming-languages synthetic-biology visualization
Last synced: 4 months ago
JSON representation
Library for combinatorially specifying, constraining, and exploring genetic design spaces.
- Host: GitHub
- URL: https://github.com/hicsail/constellation-js
- Owner: hicsail
- License: mit
- Created: 2017-11-03T16:29:27.000Z (over 7 years ago)
- Default Branch: master
- Last Pushed: 2022-12-10T16:10:03.000Z (over 2 years ago)
- Last Synced: 2024-04-24T10:53:00.559Z (about 1 year ago)
- Topics: combinatorial-search, computational-biology, constraint-programming, genetic-design, graph, graph-algorithms, javascript, programming-languages, synthetic-biology, visualization
- Language: JavaScript
- Homepage: http://constellationcad.org
- Size: 2.93 MB
- Stars: 9
- Watchers: 16
- Forks: 8
- Open Issues: 8
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# constellation-js
[](https://travis-ci.org/hicsail/constellation-js) [](https://badge.fury.io/js/constellation-js)[](https://coveralls.io/github/hicsail/constellation-js?branch=master)
### Latest stable version available at [ConstellationCAD](http://constellationcad.org/)
## Usage
### Requirements
[](https://nodejs.org/en/download/) [](https://www.python.org/downloads/)
### Local UI
```shell
git clone [email protected]:hicsail/constellation-js.git
npm run build && npm run start
```
Then open `http://localhost:8082/` on browser
### NPM Package
The package can be installed in the following way.
```shell
npm install constellation-js
```
It is possible to generate a collection of designs that match a specification using graph construction and traversals.
```javascript
const constellation = require('constellation-js');
let goldbar = '{PT7_a then galK}';
let categories = {
"PT7_a":{
"promoter": [
"PT7_WTa",
"PT7_3a",
"PT7_1a"
]
},
"galK":{
"cds": [
"galK"
]
}
}
let result = constellation.goldbar(goldbar, categories, {designName: 'my-first-design'});
// result.stateGraph, result.designs, result.sbol, result.messages
```
|Optional parameters| Description|
|--|--|
|`designName`|Name of design space for SBOL output, defaults is "constellation-design"|
|`numDesigns`|Max number of designs to enumerate, default is 20|
|`maxCycles`|Cycle depth for -orMore operators, default is 0|
|`representation`|Choose between `EDGE` or `NODE` based graph, default is EDGE|
|Output|Description|
|--|--|
|`stateGraph`|See [Graph Data Structure](#Graph-Data-Structure)|
|`designs`|List of enumerated designs|
|`sbol`| See [Synthetic Biology Open Language](#Synthetic-Biology-Open-Language)|
|`messages`| Errors or warnings, if applicable|
It is also possible to generate a collection of designs that match a specification using a purely symbolic approach (note that this approach supports only a tolerance of `0` for the AND operator and does not support the MERGE operator).
```javascript
let result = constellation.symbolic(
"(one-or-more a) then (one-or-more x)",
{"a": {"b": ["c"]}, "x":{"y": ["z1", "z2", "z3"]}},
{"numDesigns": 'all', "maxCycles":7});
```
## Case Studies
GOLDBAR syntax for the case studies described in the manuscript are available [here](demos/static/use-cases) and can be demoed on Constellation's UI via the drop down menu.
## Design Space Representations
Genetic design spaces in Constellation are represented in three ways:
1. GOLDBAR
2. Directed cyclic graph
3. SBOL
### GOLDBAR Syntax
The supported GOLDBAR concrete syntax for genetic design spaces is presented below using extended BNF notation. Notice that `then` and `.` are equivalent, and the delimiters `(`...`)` and `{`...`}` are equivalent.
```
::= then
| .
|
::= or
| and0
| and1
| and2
| merge
|
::= one-or-more
| zero-or-more
| zero-or-one
| reverse-comp
| ( )
| { }
|
::= ([A-Za-z0-9]|-|_)+
```
The JSON schema for the GOLDBAR abstract syntax tree representation (parsed from the concrete syntax presented above) can be found in [`schemas/ast.schema.json`](schemas/ast.schema.json).
### Graph Data Structure
Constellation supports both NODE and EDGE based versions of a design space. Below are examples equivalent to the GOLDBAR specification `promoter then one-or-more CDS then terminator`
|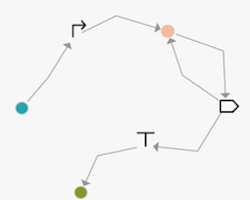 | 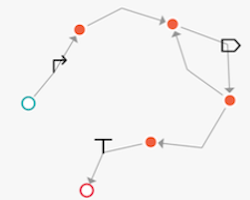|
| ------------- | ------------- |
Visualization of node-based graph| Visualization of edge-based graph
The JSON schema for a design space graph can be found in [`schemas/graph.schema.json`](schemas/graph.schema.json). Below is an example of a node-based graph within a single node in JSON format.
```
{
"604571a7-9e38-4647-afd0-9546399480b5": {
"id": "604571a7-9e38-4647-afd0-9546399480b5",
"text": "root",
"type": "root",
"edges": [
"b79407eb-95f0-4b78-99da-779f2c9cad46",
"7f6ca2fb-ef67-4687-924c-4285de896877"
],
"operator": ["One"]
}
}
```
### Synthetic Biology Open Language
[SBOL](https://sbolstandard.org/) is an open standard for the representation of *in silico* biological designs, and the icons used in this tool are provided by [SBOL Visual](https://sbolstandard.org/visual/glyphs/). Design spaces are expressed in SBOL via the `CombinatorialDerivation` extension and can be exported and stored in [Knox](https://github.com/CIDARLAB/knox). This third form of design space representation allows Constellation to be easily integrated in the synthetic biology [community](https://sbolstandard.org/applications/).