https://github.com/imagingdatacommons/cloudsegmentatorresults
https://github.com/imagingdatacommons/cloudsegmentatorresults
Last synced: 9 months ago
JSON representation
- Host: GitHub
- URL: https://github.com/imagingdatacommons/cloudsegmentatorresults
- Owner: ImagingDataCommons
- License: mit
- Created: 2024-06-03T18:20:41.000Z (over 1 year ago)
- Default Branch: main
- Last Pushed: 2024-06-12T18:56:04.000Z (over 1 year ago)
- Last Synced: 2025-02-26T18:22:28.768Z (11 months ago)
- Language: Jupyter Notebook
- Size: 1.01 MB
- Stars: 2
- Watchers: 1
- Forks: 1
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Welcome to CloudSegmentatorResults Repo
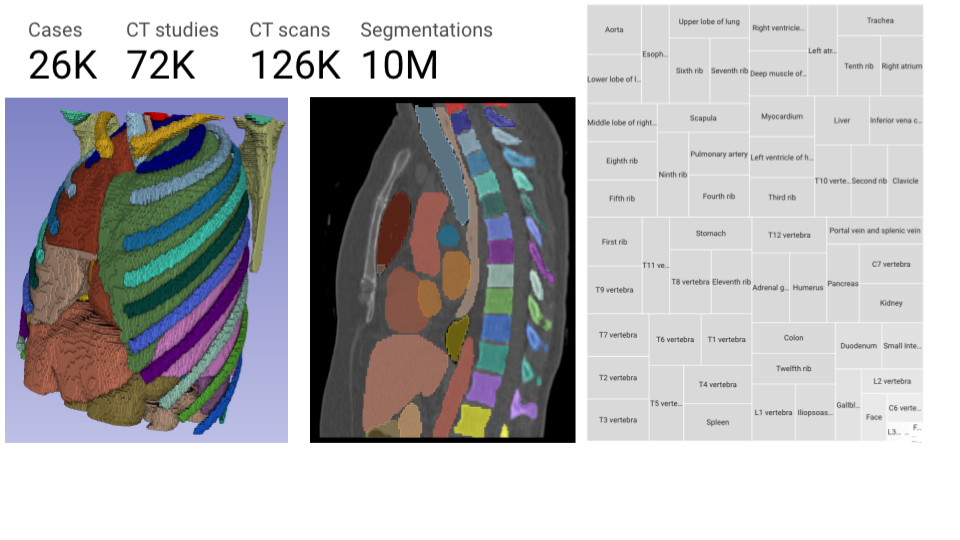
We recently made available TotalSegmentator [^1] Segmentations on NLST for over 125k CT scans with approximately 9.5 Million segmentations[^2] on Imaging Data Commons[^3]. In addition, for each segmentation, we extracted 28 shape and first-order radiomics features using pyradiomics[^4]. We encoded these Segmentations into DICOM SEG objects and shape and first-order features into DICOM Structured Reports. Moreover, we also saved pyradiomics general module features in JSON format.
This repository contains the jupyter notebooks and other related materials accompanying the following paper:
Krishnaswamy, D., Thiriveedhi, V., Ciausu, C., Clunie, D., Pieper, S., Kikinis, R. & Fedorov, A. "Rule-based outlier detection of AI-generated anatomy segmentations". Submitted to NeurIPS Datasets and Benchmarks Track 2024.
To help get a sense of the quality of these segmentations, we proposed four rule-based heuristics without the use of AI/ML.
1. Segmentation completeness: Depending on the inferior-to-superior extent of the axial CT scan, some anatomical structures may be included only partially. To remove these incomplete segmentations from further analysis, we evaluated the completeness of the segmentation by ensuring that there was at least one empty slice above and below the segment.
2. Connected component: Each anatomical region that is segmented volumetrically should be continuous and consist of a single connected component. However, using the pre-trained TotalSegmentator model with minimal post-processing can yield unconnected components for an anatomical region. Using the pyradiomics package, the VolumeNum field proves
the number of connected components, which in the ideal case should be one. We therefore used this field to identify segmentations with extraneous or noisy voxels.
3. Laterality: Segmentation algorithms may produce the incorrect laterality label (left vs right) of a region. To detect this, we evaluated the laterality by using metadata extracted from the segmentations using the pyradiomics general feature CenterOfMass field. This
attribute provides the center of mass of the region in the world-coordinate system. Using this system, the coordinates increase from right to left; therefore, we can easily determine if the laterality of a paired structure is correct.
4. Minimum volume from voxel summation: To our knowledge, the adrenal gland is the smallest organ segmented by TotalSegmentator. The average volume according to [^5] is approximately 5 mL. We used the pyradiomics feature Volume from Voxel Summation to discern volume and chose 5 mL as the thresholRule-based outlier detection of AI-generated anatomy segmentationd for all segmentations to remove artifacts.
## Interactive visualization on Hugging Face
To facilitate analysis of the segmentation results, we developed an interactive dashboard based on
the Streamlit framework (https://github.com/streamlit/streamlit) and hosted it on the
free tier of Hugging Face spaces (https://huggingface.co/spaces/ImagingDataCommons/CloudSegmentatorResults). The dashboard consists of two pages. The ‘Summary’ page contains
the results of applying each of the four heuristics to each of the segments. The table can be sorted to quickly gain insights into which of the segmentations were flagged as outliers. The "Plots" page features two types of plots and includes filtering options for radiomics features, anatomical structure, laterality, and the four heuristics. We display upset plots, which show how many segments passed or failed the heuristics, and in what combinations. Additionally, we display violin plots, which demonstrate the distributions of the standard deviation of radiomics features before and after applying the heuristics to a patient. This helps in studying the effect of the heuristics on the consistency of
a radiomics feature value distribution for a given anatomical structure within a patient. Both plots are updated dynamically depending on the choice of filters.
## Data
Two colab notebooks can be used to reproduce our work: the first focuses on downloading base tables and generating derived tables for heuristics, while the second uses the heuristics tables and explores the use cases and generates the figures presented in our manuscript.
For the [first](https://github.com/ImagingDataCommons/CloudSegmentatorResults/blob/main/part1_derivedDataGenerator.ipynb) notebook, we initially tested it on a 32vCPUs 256 GB RAM Jetstream2 instance[^6][^7].However, we made several optimizations since then to bring the RAM consumption low despite leading to a longer run times. We were able to run it successfully even on a 2vCPUs, 16 GB RAM free tier [hugging face jupyterlab space](https://huggingface.co/new-space?template=SpacesExamples%2Fjupyterlab). Run times get better if you have access to better compute resources. Other runtimes, we tested include the 2vCPU 13 GB free colab instance and the 8vCPU, 51 GB Colab Pro High-RAM instance. No GCP cloud credentials are necessary as we will be querying the metadata exported from IDC Bigquery tables as parquet files that are made available for the public for free in AWS buckets. We use duckdb, an in-memory database as it can handle highly complex data in a tiny footprint. Please check this link on how US-based researchers can request ACCESS allocations for free [^6][^7].
The [second]((https://github.com/ImagingDataCommons/CloudSegmentatorResults/blob/main/part2_exploratoryAnalysis.ipynb) notebook contains the use cases that we studied in the manuscript. We include the generation of figures that we presented in the paper, starting with the parquet files generated from the first notebook. If you want to skip the creation of the parquet files, you can jump straight to this notebook.
The following base tables, and the derived tables (and their schema) for heuristics generated by running the part 1 notebook are attached to the GitHub release (https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/tag/0.0.1):
- **[PerframeFunctionalGroupsSequence](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/nlst_totalseg_perframe.parquet)**: The DICOM SEG object contains the DICOM attribute PerframeFunctionalGroupsSequence which encodes segment and its corresponding slice locations. Due to BigQuery's limit of 1 MB per value in a cell, DICOM Segmentation Objects generated from TotalSegmentator/dcmqi that contained the DICOM attribute PerFrameFunctionalGroupsSequence over 1 MB were dropped from the BigQuery metadata table. To extract this attribute, we developed a workflow on [Terra](https://dockstore.org/my-workflows/github.com/ImagingDataCommons/CloudSegmentator/perFrameFunctionalGroupSequenceExtractionOnTerra). We unnested this attribute and are making it available to the public as a parquet file.
- **[jsonRadiomics](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/json_radiomics.parquet)**: While extracting the 28 radiomics features mentioned in the paper, we also saved the raw output from pyradiomics into a JSON file. Pyradiomics provides `general features` along with any first or shape features. We extracted and pooled the radiomics features in JSON files and provided them as a parquet file. To generate this file, we used a workflow on [Terra](https://dockstore.org/workflows/github.com/ImagingDataCommons/CloudSegmentator/convertRadiomicsJsonToDataframeOnTerra:main?tab=info), pushed the combined table to BigQuery, and then exported it as parquet files, consolidating it into one parquet file.
- **[bodyPartAndLaterality](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/bodyPartAndLaterality.parquet)**: This is an intermediate table which contains information about the body part segmented by TotalSegmentator, segment number, source CT series, and its Laterality.
- **[Segmentation Completeness](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/segmentation_completeness_table.parquet)**: This table contains info about whether a segment had at least one slice below and above the segmentation.
- **[Laterality](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/laterality_check_table.parquet)**: This table contains if laterality (left vs right) is correctly assigned by TotalSegmentator.
- **[Qualitative Checks](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/qual_checks_table.parquet)**: This table contains the three heuristics: segmentation completeness, laterality, and connected components. The fourth heuristic is added when merged with the quantitative measurements below.
- **[Flat Quantitative Measurements](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/flat_quantitative_measurements.parquet)**: This table contains the pivoted quantitative measurements for all TotalSegmentator Segmentations. Effectively each row represents a segment and all 28 radiomics features are present in their columns.
- **[qualitative_checks_and_quant_measurements](https://github.com/ImagingDataCommons/CloudSegmentatorResults/releases/download/0.0.1/qual_checks_and_quantitative_measurements.parquet)**: This is the result of combining all the heuristics along with 28 radiomics features for each segment along with the general module features VolumeNum which gives us the number of Connected Components. This file may be the most useful and is the file that is powering the Hugging Face Dashboard and all our analysis in the part 2 colab notebook.
## Acknowledgements
This work was supported in part by NIH NCI under Task Order No. HHSN2611 0071 Under Contract
No. HHSN261201500003l. Our use of JetStream2 resources was supported by the ACCESS project
230025 [^6][^7]
## References
[^1]: J Wasserthal, HC Breit, MT Meyer, M Pradella, D Hinck, AW Sauter, T Heye, DT Boll, J Cyriac, S Yang, and M Bach. Totalsegmentator: Robust segmentation of 104 anatomic structures in CT images. Radiology: Artificial Intelligence, 5:5, 2023
[^2]: VK Thiriveedhi, D Krishnaswamy, D Clunie, S Pieper, R Kikinis, and A Fedorov. Cloud-based
large-scale curation of medical imaging data using AI segmentation, 2024.
[^3]: A Fedorov, WJ Longabaugh, D Pot, DA Clunie, SD Pieper, DL Gibbs, C Bridge, MD Herrmann,
A Homeyer, R Lewis, and HJ Aerts. National cancer institute imaging data commons: Toward
transparency, reproducibility, and scalability in imaging artificial intelligence. Radiographics, 43(12):e230180, 2023
[^4]: JJ Van Griethuysen, A Fedorov, C Parmar, A Hosny, N Aucoin, V Narayan, RG Beets-Tan,
JC Fillion-Robin, S Pieper, and HJ Aerts. Computational radiomics system to decode the
radiographic phenotype. Cancer research, 77(21):e104–7, 2017
[^5]: X Wang, ZY Jin, HD Xue, W Liu, H Sun, Y Chen, and K Xu. Evaluation of normal adrenal
gland volume by 64-slice CT. Chinese Medical Sciences Journal, 27(4):220–4, 2012
[^6]: DY Hancock, J Fischer, JM Lowe, W Snapp-Childs, M Pierce, S Marru, JE Coulter, M Vaughn,
B Beck, N Merchant, and E Skidmore. Jetstream2: Accelerating cloud computing via jetstream.
In Practice and Experience in Advanced Research Computing, pages 1–8, 2021.
[^7]: TJ Boerner, S Deems, TR Furlani, SL Knuth, and J Towns. Access: Advancing innovation:
NSF’s advanced cyberinfrastructure coordination ecosystem: Services support. In Practice and
Experience in Advanced Research Computing, pages 173–176, 2023