Ecosyste.ms: Awesome
An open API service indexing awesome lists of open source software.
https://github.com/jakeret/propro
A simple pure python process profiler.
https://github.com/jakeret/propro
Last synced: about 1 month ago
JSON representation
A simple pure python process profiler.
- Host: GitHub
- URL: https://github.com/jakeret/propro
- Owner: jakeret
- License: gpl-3.0
- Created: 2016-07-26T07:38:34.000Z (over 8 years ago)
- Default Branch: master
- Last Pushed: 2024-04-30T12:48:22.000Z (8 months ago)
- Last Synced: 2024-05-02T07:38:43.346Z (8 months ago)
- Language: Python
- Homepage:
- Size: 131 KB
- Stars: 2
- Watchers: 1
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Process profiler
[](http://propro.readthedocs.io/en/latest)
A simple process profiler. **propro** can be used in many different ways. Conveniently it can be used on the command line:
```bash
$ propro --fmt=png
```
For more options, call:
```bash
$ propro --help
```
Another option is to call the profiling programmatically:
```python
import propro
x = propro.profile_cmd("ufig --background-type=chunked_map ufig.config.random")
```
The returned profiling result can then, for instance, be used for custom plotting.
**propro** offers the option to profile a single Python function using a decorator:
```python
import propro
import numpy as np
@propro.profile(sample_rate=0.1, fmt="txt")
def mem_hungry(size):
a = []
for i in range(size):
a.append(np.random.random())
b = []
for i in range(size):
t = []
for j in range(size):
t.append(i * a[j])
b.append(t)
b = np.array(b)
```
The profiling output is stored in the folder where the Python code was launched.
Finally, **propro** can be embedded in your IPython notebooks. Load the extension with:
```python
import propro
%load_ext propro
```
The profiling can be done on a line level:
```python
%propro -r 0.1 load_pixels(path, PIXEL_COUNT)
```
or on a cell level:
```python
%%propro -r 0.1
X = np.random.normal(size=(200,200,1000))
P, D, Q = np.linalg.svd(X, full_matrices=False)
X_a = np.dot(np.dot(P, np.diag(D)), Q)
print(np.std(X), np.std(X_a), np.std(X - X_a))
```
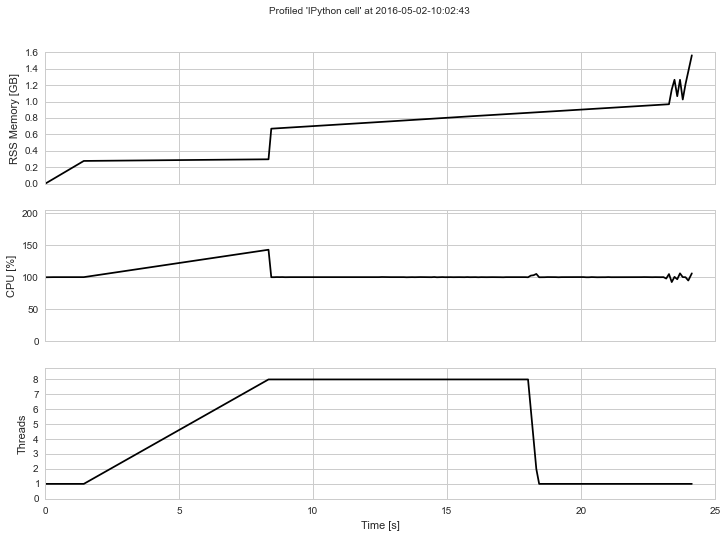
The output will look something like this if rendered into an image: