https://github.com/jenskrumsieck/porphystruct
Structural Analysis of Porphyrinoids & Corrinoids
https://github.com/jenskrumsieck/porphystruct
chemistry chemsharp conformation conformational-analysis coordination-chemistry corrinoids corrole csharp helix-toolkit macrocycle macrocycles-conformational-search nsd oxyplot porphyrinoids porphystruct simulation tetrapyrroles tu-braunschweig xray-crystallography
Last synced: 5 months ago
JSON representation
Structural Analysis of Porphyrinoids & Corrinoids
- Host: GitHub
- URL: https://github.com/jenskrumsieck/porphystruct
- Owner: JensKrumsieck
- License: mit
- Created: 2019-08-04T13:13:19.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2024-06-13T07:18:41.000Z (11 months ago)
- Last Synced: 2024-06-13T09:56:36.534Z (11 months ago)
- Topics: chemistry, chemsharp, conformation, conformational-analysis, coordination-chemistry, corrinoids, corrole, csharp, helix-toolkit, macrocycle, macrocycles-conformational-search, nsd, oxyplot, porphyrinoids, porphystruct, simulation, tetrapyrroles, tu-braunschweig, xray-crystallography
- Language: C#
- Homepage: https://porphystruct.org
- Size: 22.1 MB
- Stars: 9
- Watchers: 1
- Forks: 2
- Open Issues: 2
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README

PorphyStruct
Structual Analysis of Porphyrinoids
Download
·
Documentation
·
Report Bug
·
Statistics
[](https://codeclimate.com/github/JensKrumsieck/PorphyStruct/maintainability)
[](https://github.com/JensKrumsieck/PorphyStruct/actions/workflows/dotnet.yml)
[](https://github.com/JensKrumsieck/PorphyStruct/issues)

[](https://github.com/JensKrumsieck/PorphyStruct/releases/latest)
[](https://github.com/JensKrumsieck/PorphyStruct/releases/latest)


## Introduction
_PorphyStruct_, a new digital tool for the analysis of non-planar distortion modes of different porphyrinoids - The program makes use of the normal-coordinate structure decomposition technique (NSD) and employs sets of normal modes equivalent to those established for porphyrins in order to describe the out-of-plane dislocation pattern of perimeter atoms from corroles, norcorroles, porphycenes and other porphyrinoids quantitatively and in analogy to the established terminology.
## Table Of Contents
* [Introduction](https://github.com/JensKrumsieck/PorphyStruct#introduction)
* [Features](https://github.com/JensKrumsieck/PorphyStruct#features)
* [What is NSD?](https://github.com/JensKrumsieck/PorphyStruct#what-is-nsd)
* [Why should i care?](https://github.com/JensKrumsieck/PorphyStruct#why-should-i-care)
* [What Data is generated?](https://github.com/JensKrumsieck/PorphyStruct#what-data-is-generated)
* [How To Cite](https://github.com/JensKrumsieck/PorphyStruct#how-to-cite)
* [Further Reading](https://github.com/JensKrumsieck/PorphyStruct#further-reading)
* [System Requirements](https://github.com/JensKrumsieck/PorphyStruct#system-requirements)
* [Used Libraries](https://github.com/JensKrumsieck/PorphyStruct#used-libraries)
## Features
| | |
|-|-|
| |
| |
|
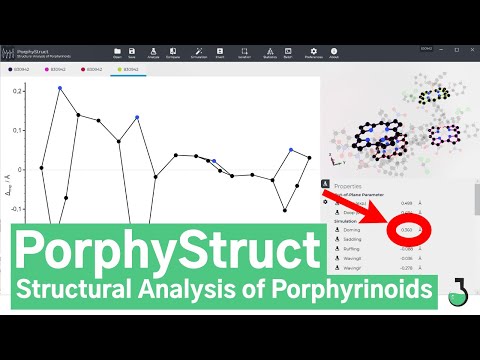
_PorphyStruct_ not only allows the generation of **displacement diagrams** (a.k.a cylinder projection) in split seconds but also offers a fast **NSD-type analysis of tetrapyrroles** with different perimeters. Additionally specific Bond distances, dihedrals oder angles such as the interplanar angle, helicity and the size of the N4 Cavity are calculated.
All perimeters derived from the following types are supported:
* **Porphyrins** (Isoporphyrins, Phtalocyanines, N-confused Porphyrins, Porphyrazines)
* **Corrole** (Isocorroles, Heterocorroles, N-confused Corroles, Corrolazines, Corrins¹)
* **Norcorrole**
* **Corrphycene**
* **Porphycene**
¹ NSD-type analysis not useful here, but diagrams are still possible.
Furthermore, the cyclic structures are automatically detected and the analysis is fully automatic for **multiple structures per crystal**. Therefore, **batch processing** and statistical evaluation is also possible.

### Features list
* Generation of Displacement Diagrams for various (**Porphyrin**, **Corrole**, **Norcorrole**, **Porphycene**, **Corrphycene** and related) tetrapyrrolic macrocycles.
* **Automatic** Recognition of the macrocylic perimeter atoms
* Analyze multiple structures per crystal (.cif, .mol2, .xyz & .pdb (1.0.1 or later) are supported see ChemSharp)
* Simulation of the generated diagrams by linear combination of standard structures based on the vibrational standard modes obtained by DFT calculations (B3LYP/def2-SVP). You can view the .XYZ Files [here](https://github.com/JensKrumsieck/PorphyStruct/tree/master/PorphyStruct.Core/Reference)
* Calculation of Bond Distances, Angles, Dihedrals, Interplanar angle, Helicity, Cavity size, ...
* Comparison of Structures and further structural analysis.
* Ablility to import and export various file types
* Mode Calculator to visualize certain combinations of vibrational modes
* Batch processing and statistics (wip)
* Automatic Update (1.0.2+)
[](https://www.youtube.com/watch?v=WdEJJrrPYKg)
## What is NSD
**NSD stands for normal-coordinate structural decomposition**. A three-dimensional molecular structure can be described by linear combinations of its vibrational modes. Shelnutt et al. were able to show in the late 1990s with the NSD method not only that all porphyrins can be described as D4h symmetric under the restrictions of the method, regardless of the substitution pattern, but also that exactly 6 vibrational modes are sufficient for the description in the vast majority of cases. These are the 6 energetically lowest vibrational modes of the porphyrin: **Doming** (A2u), **Ruffling** (B2u), **Saddling** (B1u), **Waving (X,Y)** (Eg) and **Propellering**.
_PorphyStruct_ uses a modified version of the method to make it available to other macrocyclic frameworks. The modes used are based on those of porphyrin and are not necessarily the lowest energy ones. _PorphyStruct_ provides 2 Doming, 2 Ruffling, 2 Saddling, 4 Waving and 2 Propellering modes for each type of macrocycle (Minimal Basis & Extended Basis).
### Porphyrin
|Doming|Ruffling|Saddling|Waving X|Waving Y|Propellering|
|-|-|-|-|-|-|
| |
| |
| |
| |
| |
| |
|
| |
| |
| |
| |
| |
| |
|
### Corrole
|Doming|Ruffling|Saddling|Waving X|Waving Y|Propellering|
|-|-|-|-|-|-|
| |
| |
| |
| |
| |
| |
|
| |
| |
| |
| |
| |
| |
|
### Norcorrole
|Doming|Ruffling|Saddling|Waving X|Waving Y|Propellering|
|-|-|-|-|-|-|
| |
| |
| |
| |
| |
| |
|
| |
| |
| |
| |
| |
| |
|
## Why should i care
While the conformational features of porphyrins have been extensively investigated in recent years using J.A. Shelnutt's NSD method [1], the limitations of this method have left other macrocycles such as Corrole far behind. Studies have shown correlations of conformation with photophysical and chemical properties such as reactivity. [2] For example, the metallation rates of porphyrins with large amounts of saddling are several orders of magnitude higher than for their planar counterparts. [2b]
Due to the large number of correlations found for porphyrins, a transfer of this method to other tetrapyrrolic systems is required as there a little to none correlations reported for e.g. corroles. This is the idea behind "**PorphyStruct**". PorphyStruct implements a variation of the NSD method on corroles, norcorroles, porphycenes, corrphycenes (and porphyrins).
* [1] W. Jentzen, J.-G. Ma, J. A. Shelnutt, _Biophys. J._ **1998**, _74_, 753–763; W. Jentzen, X.-Z. Song, J. A. Shelnutt, _J. Phys. Chem. B_ **1997**, _101_, 1684–1699.
* [2] a) M. O. Senge, S. A. MacGowan, J. M. O'Brien, _Chem. Commun._ **2015**, _51_, 17031–17063; M. O. Senge, _Chem. Commun._ **2006**, 243–256 b) J. Takeda, T. Ohya, M. Sato, _Inorg. Chem._ **1992**, _31_, 2877–2880.
## What Data is generated
It depends 😉
The following is available:
* Displacement diagram (as PNG or SVG).
* XY data of the deflection diagram for self-plotting (DAT or CSV)
* Analysis data (as graph (PNG/SVG), "human-readable" MD ² or "machine-readable" JSON)
* Input file converted to MOL2
* Cyclic structure converted to MOL2
* Snapshot of molecule preview
* Anything else you like to have? Tell me!
² MD Files can be opened with every Text Editor. I recommend [VS Code](https://code.visualstudio.com/).
A preview of an Analysis can be found here: [Markdown export of Structure shown below](https://github.com/JensKrumsieck/PorphyStruct/blob/master/.github/analysis/751761_analysis.md)

[Displacement Diagram](https://github.com/JensKrumsieck/PorphyStruct/blob/master/.github/analysis/751761_graph.png) and [Analysis](https://github.com/JensKrumsieck/PorphyStruct/blob/master/.github/analysis/751761_analysis.png) generated by _PorphyStruct_
**Structure**: A. B. Alemayehu, E. Gonzalez, L. K. Hansen, A. Ghosh, _Inorg. Chem._, **2009**, _48_, 7794.
## How to cite

### PorphyStruct – A Digital Tool for the Quantitative Assignment of Non-Planar Distortion Modes in Four-Membered Porphyrinoids
J. Krumsieck, M. Bröring, Chem. Eur J., 2021, 27, 11580-11588, DOI: 10.1002/chem.202101243
▶️ Very Important Paper - Front Cover - Cover Profile
#### Featured in:
* [ChemistryViews](https://www.chemistryviews.org/details/ezine/11308216/PorphyStruct_Conformational_Analysis_of_Porphyrinoids.html)
* [TU Braunschweig Magazine](https://magazin.tu-braunschweig.de/en/pi-post/digital-tools-for-observing-molecular-gymnastics/) - ([German Version](https://magazin.tu-braunschweig.de/pi-post/digitale-helfer-zur-beobachtung-von-molekuel-gymnastik/))
### Further reading
[1] W. Jentzen, J.-G. Ma, J. A. Shelnutt, _Biophys. J._ **1998**, _74_, 753–763. https://doi.org/10.1016/S0006-3495(98)74000-7
[2] W. Jentzen, X.-Z. Song, J. A. Shelnutt, _J. Phys. Chem. B_ **1997**, _101_, 1684–1699. https://doi.org/10.1021/jp963142h
[3] C. J. Kingsbury, M. O. Senge, _Coord. Chem. Rev._ **2021**, _431_, 213760. https://doi.org/10.1016/j.ccr.2020.213760
### System requirements
Windows 11 or Windows 10 (Windows 7 seems to work, too)
### Used Libraries
* [ChemSharp](https://github.com/JensKrumsieck/ChemSharp) by Jens Krumsieck (MIT License)
* [OxyPlot](https://github.com/oxyplot/oxyplot) (MIT License)
* [Math.NET Numerics](https://github.com/mathnet/mathnet-numerics) (MIT License)
* [Helix Toolkit](https://github.com/helix-toolkit/helix-toolkit) (MIT License)
* [TinyMVVM](http://github.com/JensKrumsieck/TinyMVVM) by Jens Krumsieck (MIT License)
* [ThemeCommons](http://github.com/JensKrumsieck/ThemeCommons) by Jens Krumsieck (MIT License)
* [Material Design in XAML Toolkit](https://github.com/MaterialDesignInXAML/MaterialDesignInXamlToolkit) (MIT License)