https://github.com/jeremyfix/sambiotic
https://github.com/jeremyfix/sambiotic
Last synced: 3 months ago
JSON representation
- Host: GitHub
- URL: https://github.com/jeremyfix/sambiotic
- Owner: jeremyfix
- License: mit
- Created: 2025-02-13T08:38:55.000Z (4 months ago)
- Default Branch: main
- Last Pushed: 2025-02-13T16:43:04.000Z (4 months ago)
- Last Synced: 2025-02-13T17:46:07.259Z (4 months ago)
- Language: Python
- Size: 6.84 KB
- Stars: 0
- Watchers: 1
- Forks: 0
- Open Issues: 0
-
Metadata Files:
- Readme: README.md
- License: LICENSE
Awesome Lists containing this project
README
# Examples
This gui wraps SAM2.1 for segmenting images. Examples below for
1. Segmenting diatoms
[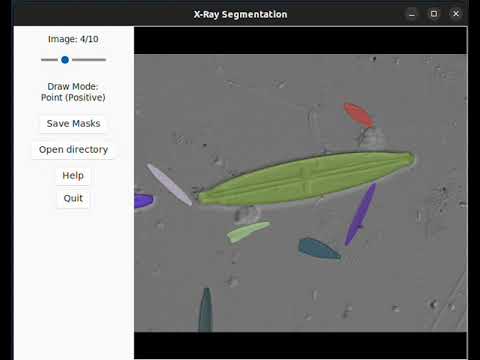](https://www.youtube.com/watch?v=PRCG1ftIQqQ)
2. Segmenting plankton
[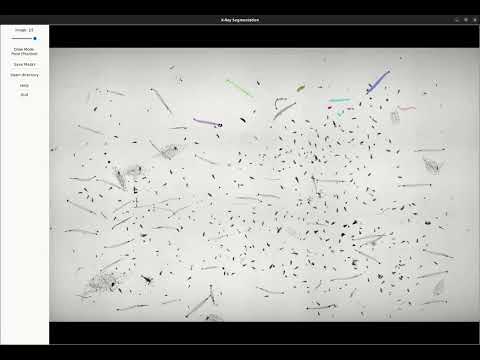](https://www.youtube.com/watch?v=vwWSzT6XBpY)
# Setup
## Install the requirements
First we install all the requirements except SAM2 which will be handled
separately.
```bash
python3 -m venv venv
source venv/bin/activate
python -m pip install -r requirements.txt
```
For installing SAM2, you could `pip install sam2`. However, it will complain at
runtime some features are missing. Therefore, we rather install it from the
sources following [the installation instructions](https://github.com/facebookresearch/sam2/blob/main/INSTALL.md).
```bash
git clone https://github.com/facebookresearch/sam2.git
python -m pip install -e "sam2[notebooks]"
```
## Download the checkpoints
For running SAM2, you need to manually download the pytorch checkpoints.
Alternatively, we could have used the ONNX exports, the installation would have
been simpler. However, as far as I know, you are then forced to resize your
images to 1024 x 1024 as expected by SAM2. Using the original basecode and the
pytorch checkpoints, we can use SAM2 on smaller images.
To get the checkpoints, meta is giving us a bash script to run :
```bash
mkdir checkpoints
cd checkpoints
wget https://raw.githubusercontent.com/facebookresearch/sam2/refs/heads/main/checkpoints/download_ckpts.sh
bash download_ckpts.sh
```
# Run
If you use a low end GPU, you may need to :
```bash
export PYTORCH_CUDA_ALLOC_CONF=expandable_segments:True
```
and then you can
```bash
python gui.py image_dir sam2.1-hiera-large
```