https://github.com/jokergoo/bsub
Send R code/R scripts/shell commands to LSF cluster without leaving R
https://github.com/jokergoo/bsub
Last synced: 6 months ago
JSON representation
Send R code/R scripts/shell commands to LSF cluster without leaving R
- Host: GitHub
- URL: https://github.com/jokergoo/bsub
- Owner: jokergoo
- License: other
- Created: 2019-11-07T22:40:28.000Z (over 5 years ago)
- Default Branch: master
- Last Pushed: 2024-06-28T10:18:53.000Z (12 months ago)
- Last Synced: 2024-12-09T21:11:05.678Z (6 months ago)
- Language: R
- Homepage: https://jokergoo.github.io/bsub
- Size: 1.51 MB
- Stars: 25
- Watchers: 3
- Forks: 5
- Open Issues: 7
-
Metadata Files:
- Readme: README.md
- Changelog: NEWS
- License: LICENSE
Awesome Lists containing this project
README
# Submitter and Monitor of the LSF Cluster
[](https://github.com/jokergoo/bsub/actions)
[](https://cran.r-project.org/web/packages/bsub/index.html)
It sends R code/R scripts/shell commands to LSF cluster without leaving R.
### Install
Directly from CRAN:
```r
install.packages("bsub")
```
Or from GitHub:
```r
if (!requireNamespace("devtools", quietly = TRUE))
install.packages("devtools")
devtools::install_github("jokergoo/bsub")
```
### Documentation
The online documentation is available at https://jokergoo.github.io/bsub/.
### Submit jobs
Directly submit R chunk:
```r
library(bsub)
# R code
bsub_chunk(name = "example", memory = 10, hours = 10, cores = 4,
{
fit = NMF::nmf(...)
# you better save `fit` into a permanent file
saveRDS(fit, file = "/path/of/fit.rds")
})
```
Submit an R script:
```r
# R script
bsub_script(name = "example",
script = "/path/of/foo.R", ...)
```
Submit shell commands:
```r
# shell commands
bsub_cmd(name = "example",
cmd = "samtools view ...", ...)
```
Kill jobs:
```r
bkill(job_id)
```
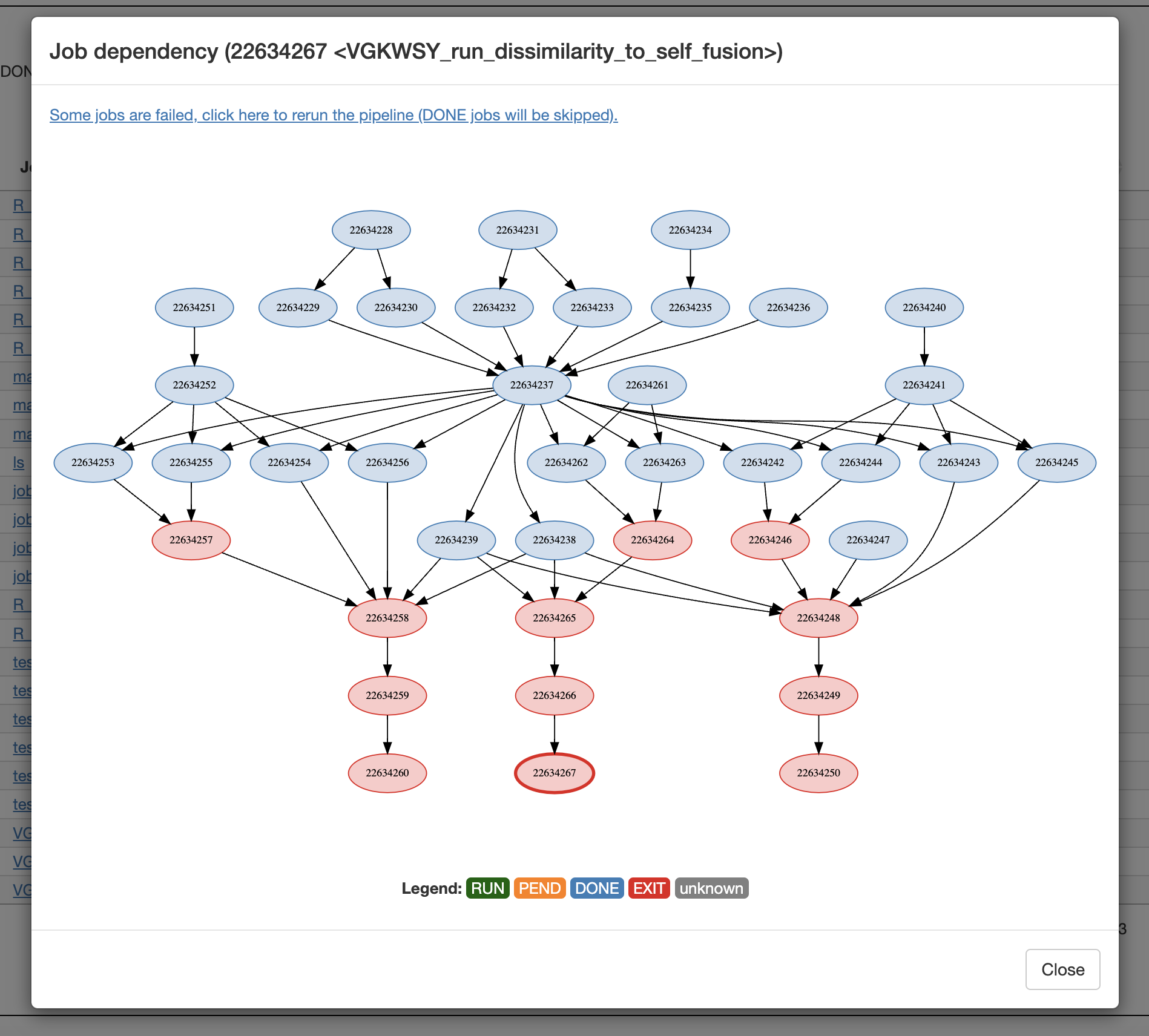
### Job dependencies
```r
job1 = bsub_chunk(...)
job2 = bsub_chunk(...)
bsub_chunk(..., dependency = c(job1, job2))
```
### View job info
View job summaries:
```r
bjobs
brecent
bjobs_running
bjobs_pending
bjobs_done
bjobs_exit
```
An example of the job queries is as follows:

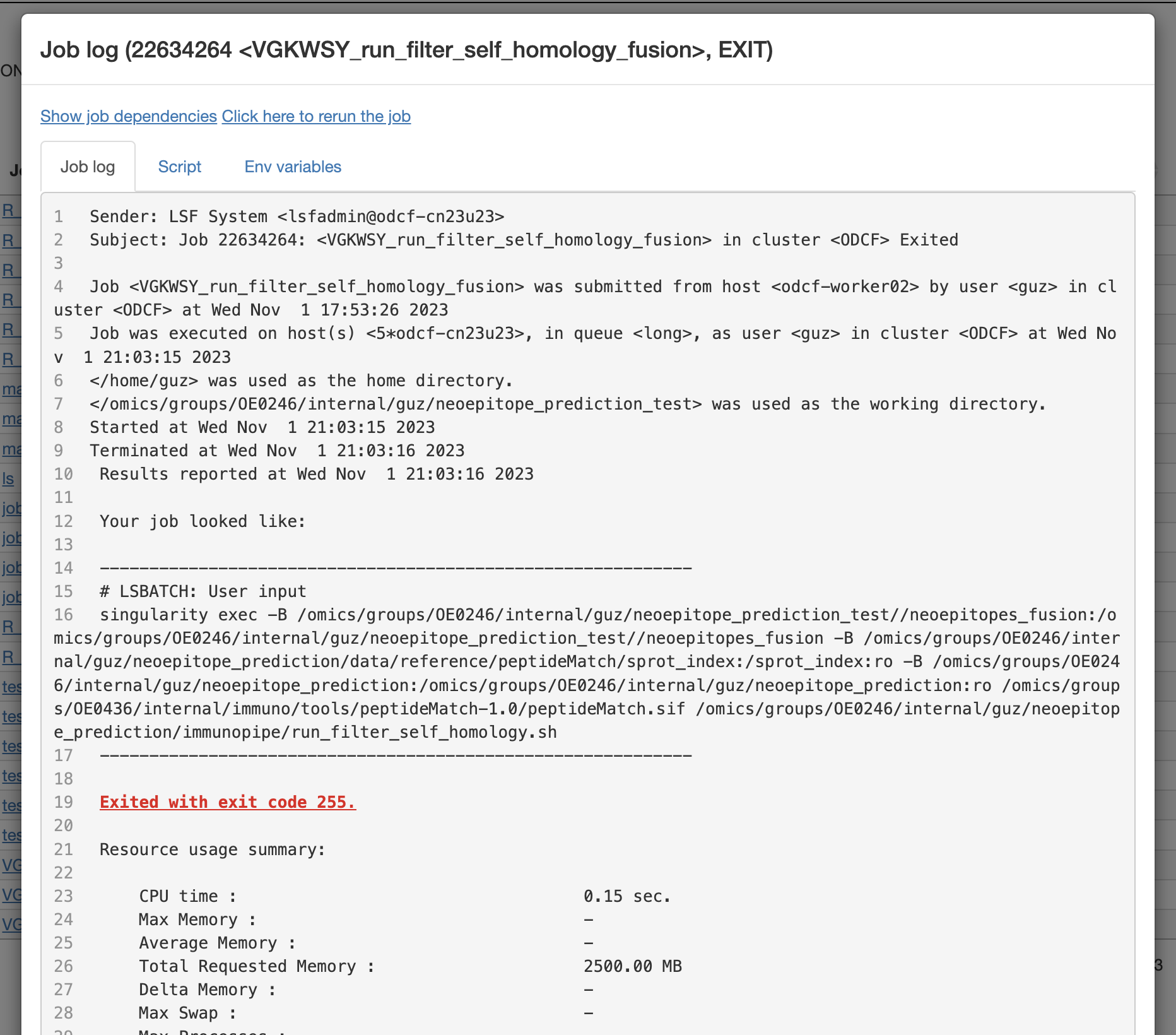
View job log:
```r
job_log(job_id)
```
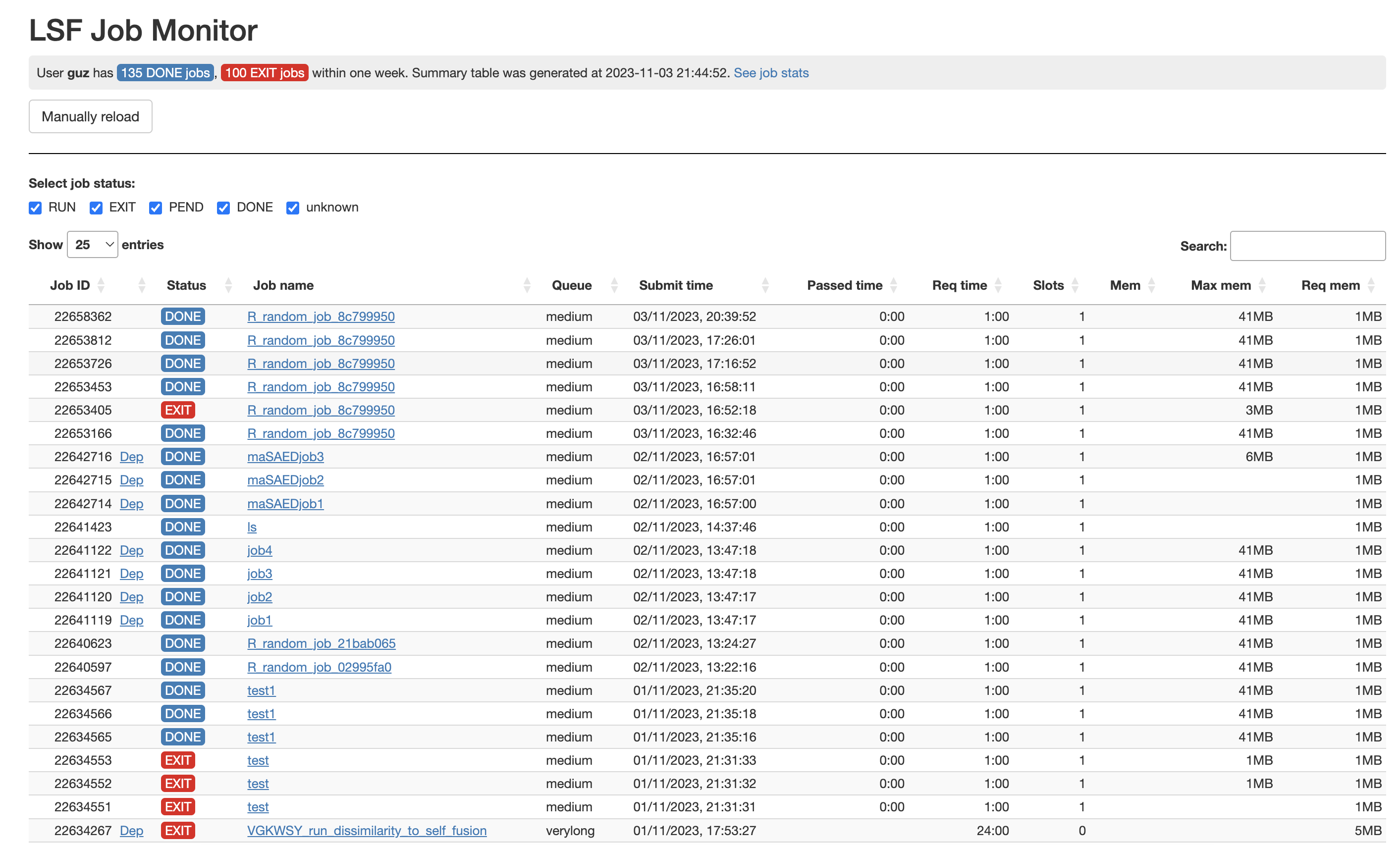
### Interactive job monitor
```r
monitor()
```
The job summary table:
Job log:
Job dependency diagram:
### License
MIT @ Zuguang Gu